Team:UC Berkeley Tools/RelatedWork
From 2008.igem.org
Related Work
This section will examine several tools which attempt to provide a design environment for synthetic biology. These can be divided into both academic and industrial offerings.
Academic
APE
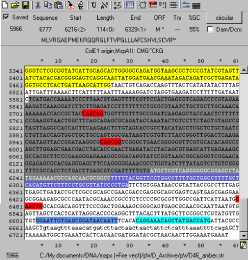
[http://www.biology.utah.edu/jorgensen/wayned/ape/ APE] (A Plasmid Editor) [6] is an editor which shows DNA sequence and translation information in a single text based viewer. It performs basic highlighting tasks for restriction sites (or user defined regions). It also shows translation, Tm, %GC, and ORF of selected DNA in real-time. It also reads DNA Strider, FASTA, Genbank and EMBL files. It has no database connectivity or notion of a larger synthetic biology community.
Athena
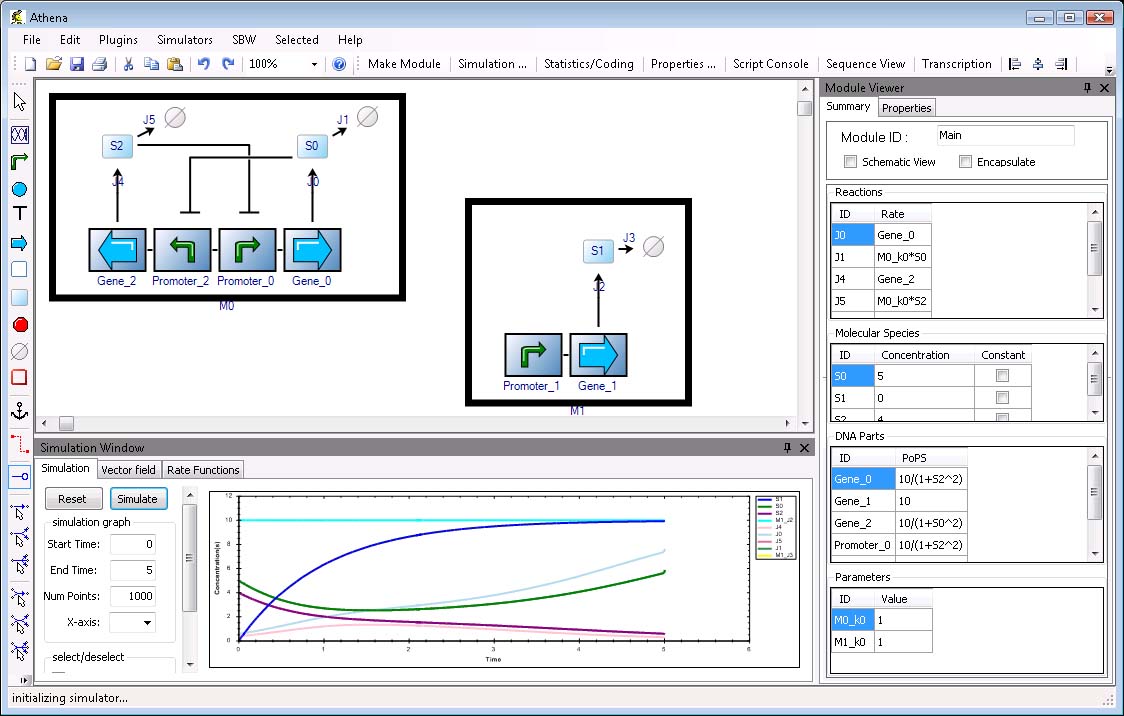
Athena is a tool developed at the University of Washington by Deepak Chandran, Frank Bergmann, Herbert Sauro. Here is their description from [http://openwetware.org/wiki/Computational_Tools OpenWetWare's Computational Tool Site]
Athena is a tool for building, simulating, and analyzing genetic circuits (as well as metabolic/signaling networks, such as SBML files). It provides a visual interface for building biological modules that can be saved and later connected together. The connection can be achieved using either the PoPS interface or by defining overlapping molecular species (similar to the concept of module in CellML and SBML). In addition to simulation, Athena supports a few other useful features: Database of Ecoli regulatory network from RegulonDB, Graphical view of part sequence, Automated derivation of transcription rate equations, Interface to all Systems Biology Workbench programs, Interface with R statistical language, and an easy plugin architecture.
You can find out more about Athena [http://www.codeplex.com/athena here].
[http://www.washington.edu/staff/deepakc/downloads/InstallAthena.exe Download Athena]
BioJADE
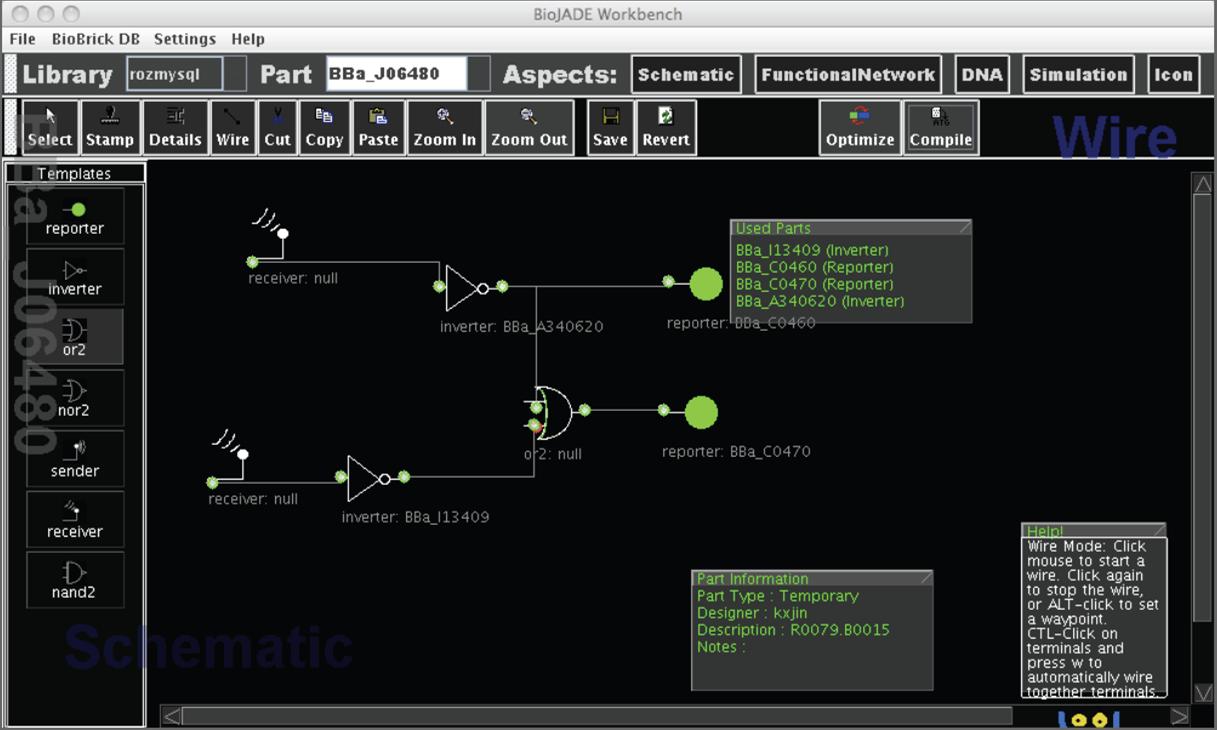
One of the early contributions in this area is [http://web.mit.edu/jagoler/www/biojade/ BioJADE] [1]. BioJADE is a design and simulation tool written in JAVA. It supports interaction with a number of part repositories including MIT's standard biological parts repository (XML interface) as well as relational database systems such as Oracle 9. It allows both the importing of devices from repositories as well as the submission of newly created parts to these same repositories. In terms of manipulating the design, BioJADE provides four aspects. These are Schematic, DNA, Functional, and Icon. As shown, Schematic presents a traditional schematic editor view for the design of logic circuits. The DNA aspect illustrates the actual DNA nucleotide sequence of the design. Functional Network presents a closer view of the layout of the DNA. It is similar to the DNA aspect but allows the user to edit the sequence. The Icon aspect allows the designer to create icons for parts to be used in the schematic aspect. Input, output, and undefined terminals are specified either as a static number or as dynamic (added at runtime). Finally, BioJADE offers a simulation mode where the design can be simulated. It interacts with simulators like Stochastirator [3] or Tabasco [4].
BioStudio
BioStudio is the work of researchers at Johns Hopkins University (Joel Bader, Jef Boeke, and Sarah Richardson).
This is taken from BioStudio press release material at [http://research.microsoft.com/ur/us/fundingopps/RFPs/Synthetic_Biology_Awards_2006.aspx Computational Challenges in Synthetic Biology 2006 Awards]
We propose BioStudio as a collaborative integrated design environment (IDE) providing editing and revision control for synthetic genomes. Windows SharePoint Services and SQL Server will provide back-end support. The server middle-layer will extend existing open-source visualization-only software (GBrowse) with editing functions, including a BioBricks palette, and will communicate with the back-end using .NET and FrontPage Remote Procedure Calls. Genome sequence and annotations will be stored in XML with an adapter to the Generic Feature Format (GFF) standard. Synthetic biology teams will connect with a web browser client. Development and testing will proceed in conjunction with an in-house project to engineer a synthetic yeast cell that encodes a combinatorial satisfiability experiment to identify a minimal genome. Our physical-level editor is complementary to logical-level design tools such as BioJADE. It provides a multiscale visualization metaphor that enables non-computer-scientist teams to collaborate on documents that are too large for "Track changes" and too critical or interdependent to be consigned to a Wiki. Software will be released under a BSD license.
In addition to BioStudio there is an interactive web site called [http://www.genedesign.org Gene Design].
GenoCAD
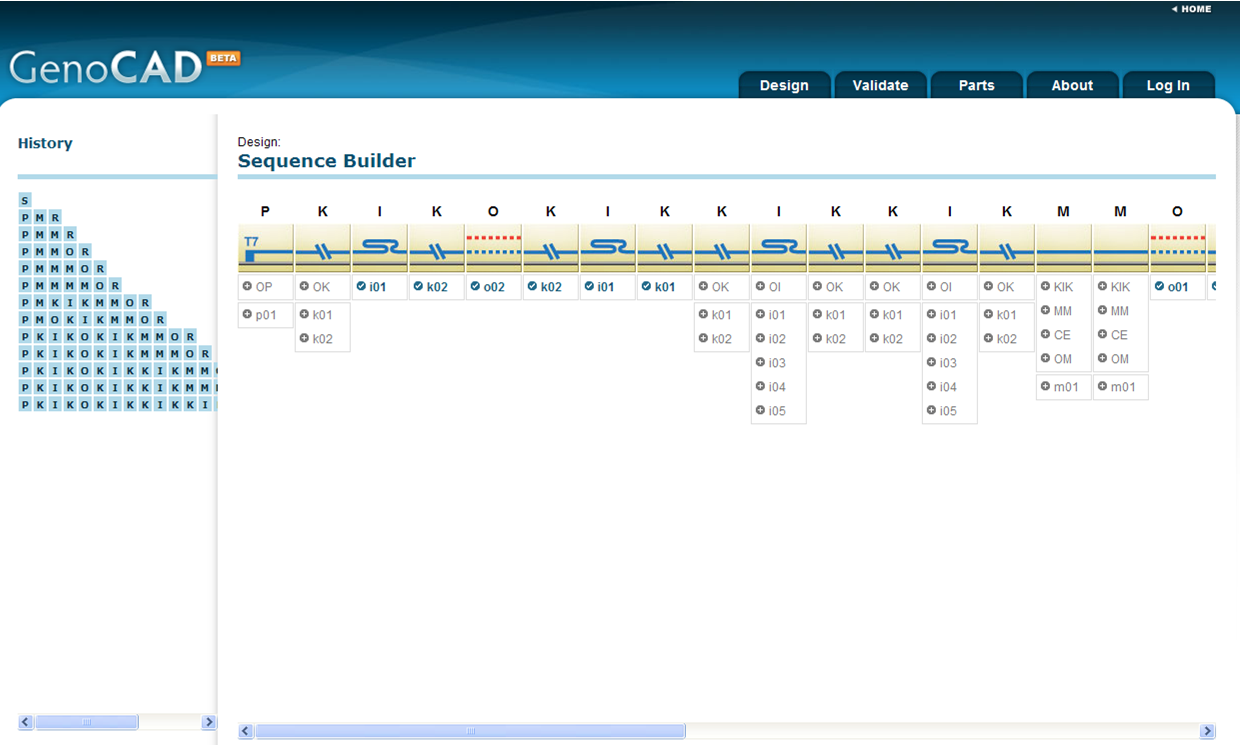
[http://synbio.vbi.vt.edu:25500/genocad/ GenoCAD] [5] is a web based tool which guides you through the process of creating new genetic constructs by combining DNA sequences known as genetic parts or biological parts. Users can create their own account on the web site to add their new genetic parts, customize the build-in parts libraries for their own needs, and save their designs for future reference. Upon completion of the design process, the design DNA sequence can be saved as a text file for synthesis by a gene synthesis company or simply for further analysis in sequence analysis applications. GenoCAD also provides a construct validation feature (using a context free grammar (CFG) analysis) that allows users to import the sequence of a design and check its validity. GenoCAD relies on a linguistic model of the DNA sequence. The site currently supports a single proof of concept grammar that has been described in a paper published in Bioinformatics. Ongoing development efforts aim at implementing support for multiple grammars in GenoCAD.
Strengths of this tool are primarily its attempt to validate the sequence and categorize parts via production rules based on 26 syntactic categories (i.e. gene coding region, terminator, stop codon, promoter, ribosome binding site, etc).
For more information see: [http://bioinformatics.oxfordjournals.org/cgi/reprint/btm446v2.pdf A syntactic model to design and verify synthetic genetic constructs derived from standard biological parts]
Industrial
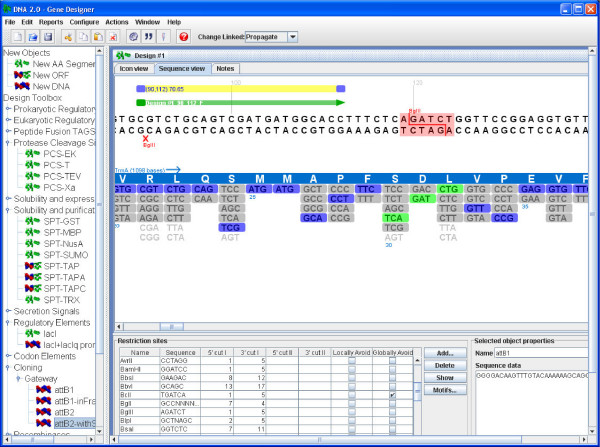
Gene Designer
[http://www.dna20.com/tools/genedesigner.php Gene Designer] [2] by DNA 2.0 is a stand alone graphical software package for manipulating biological designs. The software enables you to combine building blocks such as regulatory DNA elements, amino acid sequences, affinity tags and cloning features. Additionally you can optimize expression by codon optimizing proteins for any expression host. There are two design views for the user, Icon and Sequence. The icon view encapsulates the DNA sequence as a specific icon indicating what type of part it is. The sequence view is an annotated DNA sequence corresponding to the Icon view. Gene Designer supports a wide variety of computational manipulations to the design such as codon optimization and can identify various DNA regions. Designs can be imported/exported in FASTA format for use with other tools and flows. There is not currently support for external databases (but is forthcoming in version 2.0).
Here is a list of features as given by DNA 2.0:
- Manage genetic elements, constructs and libraries of constructs via drag and drop
- Create/Edit/Split/Merge/Save genetic elements (AA and DNA sequences)
- Back-translate open reading frames using a codon bias table
- Add/Remove restriction sites or other motifs (locally or globally)
- Design sequencing primers for a construct, view sequencing trace files aligned construct [**]
- Perform /in silico/ plasmid cloning, show circular plasmid map [**]
- Annotate and Measure (TM, GC%, etc.) construct regions [**]
- Create, Download, Edit & Merge codon bias tables [**]
- Support user specific operations via scripting environment [**]
- Import and Export various file formats Fasta, GenBank, etc. [**]
- E-commerce enabled
References
- J. Goler. BioJADE: A Design and Simulation Tool for Synthetic Biological Systems. Master’s thesis, MIT, MIT Computer Science and Artificial Intelligence Laboratory, May 2004.
- A. Villalobos, J. E. Ness, C. Gustafsson, J. Minshull, and S. Govindarajan. Gene designer: a synthetic biology tool for constructing artificial dna segments. BMC Bioinformatics, 7:285, June 2006.
- D. T. Gillespie. A general method for numerically simulating the stochastic time evolution of coupled chemical reactions. Journal of Computational Physics, 22(4):403–434, December 1976.
- S. Kosuri, J. Kelly, and D. Endy. Tabasco: A single molecule, base-pair resolved gene expression simulator. BMC Bioinformatics, 8:480, December 2007.
- Y. Cai, B. Hartnett, C. Gustafsson, and J. Peccoud. A syntactic model to design and verify synthetic genetic constructs derived from standard biological parts. Bioinformatics, 23(20):2760–2767, October 2007.
- M. W. Davis. A Plasmid Editor. World Wide Web, http://www.biology.utah.edu/jorgensen/wayned/ape/, 2008.
[**]Partially implemented at present, planned for version 2 release
 "
"