Team:Heidelberg/Modeling
From 2008.igem.org
(→Our achievements) |
|||
| (90 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | + | __NOTOC__ | |
| - | + | <html> | |
| - | |||
| - | |||
| - | |||
| - | + | <style> | |
| - | + | h1.firstHeading { display: none; } | |
| - | + | ||
| - | + | ||
| - | + | p {text-align: justify;} | |
| - | { | + | a:link { color: #00b0e6; text-decoration: none} |
| - | + | a:visited { color:#00b0e6; text-decoration: none} | |
| - | + | a:hover { color:#f29400; text-decoration: none} | |
| - | + | a:active { color:#f29400; text-decoration: none} | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | #bodyContent { padding: 10px auto; width: 910px; margin: auto; clear: none; } | ||
| + | table#team_members { text-align: justify; border: 0; } | ||
| + | table#team_members h2, table#team_members h3 { clear: both; } | ||
| - | |||
| - | + | /*-----------------------------------------------------------------------------------------------*/ | |
| + | div.MenuBar ul li ul.DropDownMenu { | ||
| + | display: none; /* Hides all drop-down menus. */ | ||
| + | |||
| + | } | ||
| + | /* | ||
| + | li:hover works in IE7 and FF2. | ||
| + | a:hover works in IE6 and FF2. | ||
| + | a:hover breaks li:hover in FF2. | ||
| + | */ | ||
| + | div.MenuBar ul li:hover ul.DropDownMenu li ul.SideMenu, | ||
| + | div.MenuBar ul li a:hover ul.DropDownMenu li a ul.SideMenu { | ||
| + | display: none; /* Hides all side menus. */ | ||
| + | } | ||
| + | /*------------------------------------------------------------------------------------- Menu Bar */ | ||
| + | div.MenuBar { | ||
| + | font: arial, helvetica, sans-serif; | ||
| + | height: 30px; | ||
| + | width: 910px; | ||
| + | /*width: 100%*/ | ||
| + | margin: 0; | ||
| + | border-top: 0; | ||
| + | border-right: 0; | ||
| + | border-left: 0; | ||
| + | padding: 0; | ||
| + | background: black; | ||
| + | |||
| + | } | ||
| + | div.MenuBar ul { | ||
| + | font: arial, helvetica, sans-serif; | ||
| + | text-align: center; | ||
| + | list-style-type: none; | ||
| + | margin: 0.5em auto; | ||
| + | border: 0; | ||
| + | padding: 0; | ||
| + | background: black; | ||
| + | } | ||
| + | div.MenuBar ul li { | ||
| + | font: arial, helvetica, sans-serif; | ||
| + | display: block; | ||
| + | padding: 0; | ||
| + | margin: 0; | ||
| + | font-size: 1.3em; | ||
| + | float: left; | ||
| + | background: black; | ||
| + | text-align: center; | ||
| + | width: 107px; | ||
| + | position: relative; /* Sets the positioning context for each drop-down menu. */ | ||
| + | } | ||
| + | |||
| + | div.MenuBar ul li a { | ||
| + | font: arial, helvetica, sans-serif; | ||
| + | display: block; | ||
| + | background: black; | ||
| + | height: 22px; /* Keep height + padding-top + padding-bottom sync with the menu bar height. */ | ||
| + | color: #ffffff; | ||
| + | padding-top: 4px; | ||
| + | padding-bottom: 4px; | ||
| + | padding-left: 1em; /* Sets the left space between top-level items. */ | ||
| + | padding-right: 1em; /* Sets the right space between top-level items. */ | ||
| + | text-decoration: none; | ||
| + | } | ||
| + | |||
| + | /*------------------------------------------------------------------------------ Drop-Down Menus */ | ||
| + | div.MenuBar ul li:hover ul.DropDownMenu, | ||
| + | div.MenuBar ul li a:hover ul.DropDownMenu { | ||
| + | display: block; | ||
| + | width: 10em; /* Drop-down menu width. | ||
| + | Use MenuTailor.css to customize. */ | ||
| + | height: 1em; | ||
| + | padding: 1px; /* Sets the drop-down menu "effective border" width. */ | ||
| + | position: absolute; | ||
| + | top: 23px; /* Places the drop-down menu under the menu bar. | ||
| + | Keep it sync with the menu bar height. */ | ||
| + | left: 0; /* Aligns the drop-down menu to its top-level item. */ | ||
| + | background-color: black; /* Selected item. */ | ||
| + | color: #FFFFFF; | ||
| + | |||
| + | } | ||
| + | div.MenuBar ul li:hover ul.DropDownMenu li a, | ||
| + | div.MenuBar ul li a:hover ul.DropDownMenu li a { | ||
| + | width: 10em; /* Keep it sync with the drop-down menu width. | ||
| + | Use MenuTailor.css to customize. */ | ||
| + | height: 1em; | ||
| + | padding-left: 0; | ||
| + | padding-right: 0; | ||
| + | background-color: black; /* Selected item. */ | ||
| + | color: #FFFFFF; | ||
| + | } | ||
| + | ul.DropDownMenu li a span { | ||
| + | display: block; | ||
| + | padding-left: 0.75em; /* Sets the left space of each drop-down menu item. */ | ||
| + | padding-right: 0.25em; /* Sets the right space of each drop-down menu item. */ | ||
| + | text-align: right; /* Aligns the >> symbol to the right. */ | ||
| + | } | ||
| + | ul.DropDownMenu li a span span { | ||
| + | float: left; /* Aligns the text (back) to the left. */ | ||
| + | font: 12px arial, helvetica, sans-serif; /* Required for IE55. */ | ||
| + | height: 20px; | ||
| + | color: #FFFFFF; | ||
| + | } | ||
| + | /*----------------------------------------------------------------------------------- Side Menus */ | ||
| + | div.MenuBar ul li:hover ul.DropDownMenu li:hover ul.SideMenu, | ||
| + | div.MenuBar ul li a:hover ul.DropDownMenu li a:hover ul.SideMenu { | ||
| + | display: block; | ||
| + | width: 11em; /* Side menu width. | ||
| + | Use MenuTailor.css to customize. */ | ||
| + | padding: 1px; /* Sets the side menu "effective border" width. */ | ||
| + | position: absolute; | ||
| + | top: -1px; /* Aligns the side menu to its drop-down menu item. | ||
| + | Keep it sync with the side menu "effective border" width. */ | ||
| + | left: 13em; /* Places the side menu to the right of the drop-down menu. | ||
| + | Keep it sync with the drop-down menu width. | ||
| + | Use MenuTailor.css to customize. */ | ||
| + | } | ||
| + | div.MenuBar ul li:hover ul.DropDownMenu li:hover ul.SideMenu li a, | ||
| + | div.MenuBar ul li a:hover ul.DropDownMenu li a:hover ul.SideMenu li a { | ||
| + | width: 11em; /* Keep it sync with the side menu width. | ||
| + | Use MenuTailor.css to customize. */ | ||
| + | font: 12px arial, helvetica, sans-serif; /* Required for IE55. */ | ||
| + | left: 13em; /* Places the side menu to the right of the drop-down menu. | ||
| + | Keep it sync with the drop-down menu width. | ||
| + | Use MenuTailor.css to customize. */ | ||
| + | } | ||
| + | div.MenuBar ul li ul.DropDownMenu li ul.SideMenu li a span { | ||
| + | padding-left: 1.5em; /* Sets the left space of each side menu item. */ | ||
| + | padding-right: 0.5em; /* Sets the right space of each side menu item. */ | ||
| + | text-align: left; | ||
| + | font: 12px arial, helvetica, sans-serif; /* Required for IE55. */ | ||
| + | left: 13em; /* Places the side menu to the right of the drop-down menu. | ||
| + | Keep it sync with the drop-down menu width. | ||
| + | Use MenuTailor.css to customize. */ | ||
| + | } | ||
| + | /*----------------------------------------------------------------------------- Browser Specific */ | ||
| + | * html div.MenuBar ul li a { | ||
| + | float: left; /* Required for IE55 and IE6. | ||
| + | Breaks O9. | ||
| + | Hidden (* html) from non-IE browsers. */ | ||
| + | } | ||
| + | * html ul.DropDownMenu li a:hover { | ||
| + | cursor: hand; /* Required for IE55. | ||
| + | Hidden (* html) from non-IE browsers. */ | ||
| + | } | ||
| + | ul.DropDownMenu li a:hover { | ||
| + | cursor: pointer; /* Required for IE6 and IE7. | ||
| + | Hidding it (* html) from non-IE browsers breaks IE7! | ||
| + | } | ||
| + | * html div.MenuBar a:hover { | ||
| + | text-decoration: none; /* Required for IE55 and IE6. | ||
| + | Hidden (* html) from non-IE browsers. */ | ||
| + | } | ||
| + | * html div.MenuBar ul li table, | ||
| + | * html div.MenuBar ul li table td { | ||
| + | border: 0; /* Required for IE55 and IE6. | ||
| + | Hidden (* html) from non-IE browsers. */ | ||
| + | } | ||
| + | /*------------------------------------------------------------------------------- Default Colors */ | ||
| + | div.MenuBar { | ||
| + | background-color: Menu; | ||
| + | border-bottom: 1px solid ButtonShadow; | ||
| + | } | ||
| + | div.MenuBar a { | ||
| + | background-color: Menu; /* Top-level unselected items. */ | ||
| + | color: MenuText; | ||
| + | } | ||
| + | div.MenuBar ul li:hover a, | ||
| + | div.MenuBar ul li a:hover { | ||
| + | color: #ea7f16; | ||
| + | background-color: Highlight; /* Top-level selected item. */ | ||
| + | color: HighlightText; | ||
| + | } | ||
| + | /*...............................................................................................*/ | ||
| + | div.MenuBar ul li:hover ul.DropDownMenu, | ||
| + | div.MenuBar ul li a:hover ul.DropDownMenu { | ||
| + | background-color: ButtonShadow; /* Sets the drop-down menu "effective border" color. */ | ||
| + | } | ||
| + | div.MenuBar ul li:hover ul.DropDownMenu li a, | ||
| + | div.MenuBar ul li a:hover ul.DropDownMenu li a { | ||
| + | background-color: Menu; Drop-down menu unselected items. | ||
| + | Sets the drop-down menu "effective background" color. */ | ||
| + | color: MenuText; | ||
| + | } | ||
| + | div.MenuBar ul li:hover ul.DropDownMenu li:hover a, | ||
| + | div.MenuBar ul li a:hover ul.DropDownMenu li a:hover { | ||
| + | background-color: Highlight; /* Drop-down menu selected item. */ | ||
| + | color: HighlightText; | ||
| + | } | ||
| + | /*...............................................................................................*/ | ||
| + | div.MenuBar ul li:hover ul.DropDownMenu li:hover ul.SideMenu, | ||
| + | div.MenuBar ul li a:hover ul.DropDownMenu li a:hover ul.SideMenu { | ||
| + | background-color: ButtonShadow; /* Sets the side menu "effective border" color. */ | ||
| + | } | ||
| + | div.MenuBar ul li:hover ul.DropDownMenu li:hover ul.SideMenu li a, | ||
| + | div.MenuBar ul li a:hover ul.DropDownMenu li a:hover ul.SideMenu li a { | ||
| + | background-color: Menu; /* Side menu unselected items. | ||
| + | Sets the side menu "effective background" color. */ | ||
| + | color: MenuText; | ||
| + | } | ||
| + | div.MenuBar ul li:hover ul.DropDownMenu li:hover ul.SideMenu li a:hover, | ||
| + | div.MenuBar ul li a:hover ul.DropDownMenu li a:hover ul.SideMenu li a:hover { | ||
| + | background-color: Highlight; /* Side menu selected item. */ | ||
| + | color: HighlightText; | ||
| + | } | ||
| + | /*-----------------------------------------------------------------------------------------------*/ | ||
| + | /*Script-Free 3-Level Menu 1.2 Tailor | ||
| + | www.CesarDaniel.info | ||
| + | /*-------------------------------------------------------------------------------------- General */ | ||
| + | body { | ||
| + | background: white; | ||
| + | color: black; | ||
| + | margin: 0; | ||
| + | border: 0; | ||
| + | padding: 0; | ||
| + | } | ||
| + | |||
| + | |||
| + | div.MenuBar { | ||
| + | font: 13px arial, helvetica, sans-serif; | ||
| + | } | ||
| + | div.MenuBar ul { | ||
| + | font: 13px arial, helvetica, sans-serif; /* Required for IE55. */ | ||
| + | } | ||
| + | /*--------------------------------------------------------------------------------------- Colors */ | ||
| + | div.MenuBar { | ||
| + | background-color: black; /* Selected item. */ | ||
| + | color: #FFFFFF; | ||
| + | border-bottom: 1px solid ButtonShadow; | ||
| + | } | ||
| + | div.MenuBar a, | ||
| + | div.MenuBar ul li:hover ul.DropDownMenu li a, | ||
| + | div.MenuBar ul li a:hover ul.DropDownMenu li a, | ||
| + | div.MenuBar ul li:hover ul.DropDownMenu li:hover ul.SideMenu li a, | ||
| + | div.MenuBar ul li a:hover ul.DropDownMenu li a:hover ul.SideMenu li a { | ||
| + | background-color: black; /* Selected item. */ | ||
| + | color: #FFFFFF; | ||
| + | } | ||
| + | div.MenuBar ul li:hover a, | ||
| + | div.MenuBar ul li a:hover, | ||
| + | div.MenuBar ul li:hover ul.DropDownMenu li:hover a, | ||
| + | div.MenuBar ul li a:hover ul.DropDownMenu li a:hover, | ||
| + | div.MenuBar ul li:hover ul.DropDownMenu li:hover ul.SideMenu li a:hover, | ||
| + | div.MenuBar ul li a:hover ul.DropDownMenu li a:hover ul.SideMenu li a:hover { | ||
| + | background-color: #00b0e6; /* Selected item. */ | ||
| + | color: #FFFFFF; | ||
| + | } | ||
| + | div.MenuBar ul li:hover ul.DropDownMenu, | ||
| + | div.MenuBar ul li a:hover ul.DropDownMenu, | ||
| + | div.MenuBar ul li:hover ul.DropDownMenu li:hover ul.SideMenu, | ||
| + | div.MenuBar ul li a:hover ul.DropDownMenu li a:hover ul.SideMenu { | ||
| + | background-color: ButtonShadow; /* Sets the drop-down and side menus "effective border" color. */ | ||
| + | } | ||
| + | /*--------------------------------------------------------------------------------------- Widths */ | ||
| + | /* | ||
| + | |||
| + | /* | ||
| + | Menu Bar 1 | ||
| + | Drop-Down Menu #2 | ||
| + | */ | ||
| + | div.MenuBar#navi ul li:hover ul.DropDownMenu#MB1-DDM4, | ||
| + | div.MenuBar#navi ul li a:hover ul.DropDownMenu#MB1-DDM4, | ||
| + | div.MenuBar#navi ul li:hover ul.DropDownMenu#MB1-DDM4 li a, | ||
| + | div.MenuBar#navi ul li a:hover ul.DropDownMenu#MB1-DDM4 li a { | ||
| + | width: 11em; /* Drop-down menu width. */ | ||
| + | } | ||
| + | div.MenuBar#navi ul li:hover ul.DropDownMenu#MB1-DDM5, | ||
| + | div.MenuBar#navi ul li a:hover ul.DropDownMenu#MB1-DDM5, | ||
| + | div.MenuBar#navi ul li:hover ul.DropDownMenu#MB1-DDM5 li a, | ||
| + | div.MenuBar#navi ul li a:hover ul.DropDownMenu#MB1-DDM5 li a { | ||
| + | width: 12em; /* Drop-down menu width. */ | ||
| + | } | ||
| + | |||
| + | /*...............................................................................................*/ | ||
| + | /* | ||
| + | Menu Bar 1 | ||
| + | Drop-Down Menu #2 | ||
| + | Side Menu #1 | ||
| + | */ | ||
| + | div.MenuBar#navi ul li:hover ul.DropDownMenu li:hover ul.SideMenu#MB1-DDM2-SM1, | ||
| + | div.MenuBar#navi ul li a:hover ul.DropDownMenu li a:hover ul.SideMenu#MB1-DDM2-SM1 { | ||
| + | left: 15.5em !important; /* Places the side menu to the right of the drop-down menu. | ||
| + | Keep it sync with the drop-down menu width. */ | ||
| + | } | ||
| + | div.MenuBar#navi ul li:hover ul.DropDownMenu li:hover ul.SideMenu#MB1-DDM2-SM1, | ||
| + | div.MenuBar#navi ul li a:hover ul.DropDownMenu li a:hover ul.SideMenu#MB1-DDM2-SM1, | ||
| + | div.MenuBar#navi ul li:hover ul.DropDownMenu li:hover ul.SideMenu#MB1-DDM2-SM1 li a, | ||
| + | div.MenuBar#navi ul li a:hover ul.DropDownMenu li a:hover ul.SideMenu#MB1-DDM2-SM1 li a { | ||
| + | width: 10em; /* Side menu width. */ | ||
| + | } | ||
| + | /*...............................................................................................*/ | ||
| + | /* | ||
| + | Menu Bar 1 | ||
| + | Drop-Down Menu #2 | ||
| + | Side Menu #2 | ||
| + | */ | ||
| + | div.MenuBar#navi ul li:hover ul.DropDownMenu li:hover ul.SideMenu#MB1-DDM2-SM2, | ||
| + | div.MenuBar#navi ul li a:hover ul.DropDownMenu li a:hover ul.SideMenu#MB1-DDM2-SM2 { | ||
| + | left: 15.5em !important; /* Places the side menu to the right of the drop-down menu. | ||
| + | Keep it sync with the drop-down menu width. */ | ||
| + | } | ||
| + | div.MenuBar#navi ul li:hover ul.DropDownMenu li:hover ul.SideMenu#MB1-DDM2-SM2, | ||
| + | div.MenuBar#navi ul li a:hover ul.DropDownMenu li a:hover ul.SideMenu#MB1-DDM2-SM2, | ||
| + | div.MenuBar#navi ul li:hover ul.DropDownMenu li:hover ul.SideMenu#MB1-DDM2-SM2 li a, | ||
| + | div.MenuBar#navi ul li a:hover ul.DropDownMenu li a:hover ul.SideMenu#MB1-DDM2-SM2 li a { | ||
| + | width: 10em; /* Side menu width. */ | ||
| + | } | ||
| + | /*...............................................................................................*/ | ||
| + | /* | ||
| + | Menu Bar 1 | ||
| + | Drop-Down Menu #2 | ||
| + | Side Menu #3 | ||
| + | */ | ||
| + | div.MenuBar#navi ul li:hover ul.DropDownMenu li:hover ul.SideMenu#MB1-DDM2-SM3, | ||
| + | div.MenuBar#navi ul li a:hover ul.DropDownMenu li a:hover ul.SideMenu#MB1-DDM2-SM3 { | ||
| + | left: 15.5em !important; /* Places the side menu to the right of the drop-down menu. | ||
| + | Keep it sync with the drop-down menu width. */ | ||
| + | } | ||
| + | div.MenuBar#navi ul li:hover ul.DropDownMenu li:hover ul.SideMenu#MB1-DDM2-SM3, | ||
| + | div.MenuBar#navi ul li a:hover ul.DropDownMenu li a:hover ul.SideMenu#MB1-DDM2-SM3, | ||
| + | div.MenuBar#navi ul li:hover ul.DropDownMenu li:hover ul.SideMenu#MB1-DDM2-SM3 li a, | ||
| + | div.MenuBar#navi ul li a:hover ul.DropDownMenu li a:hover ul.SideMenu#MB1-DDM2-SM3 li a { | ||
| + | width: 10em; /* Side menu width. */ | ||
| + | } | ||
| + | /*...............................................................................................*/ | ||
| + | |||
| + | </style> | ||
| + | |||
| + | |||
| + | <body> | ||
| + | <div id="header"><img src="https://static.igem.org/mediawiki/2008/7/78/TeamHeidelbergHeader.jpg" alt="Team Heidelberg" /></div> | ||
| + | <div class='MenuBar' id="navi"> | ||
| + | <ul> | ||
| + | <li> | ||
| + | <a href="https://2008.igem.org/Team:Heidelberg" style="color: white">Home | ||
| + | <!--[if gt IE 6]><!--></a><!--<![endif]--> | ||
| + | <!--[if lt IE 7]><table border="0" cellpadding="0" cellspacing="0"><tr><td><![endif]--> | ||
| + | <!--[if lte IE 6]></td></tr></table></a><![endif]--> | ||
| + | </li> | ||
| + | <li> | ||
| + | <a href="https://2008.igem.org/Team:Heidelberg/Team" style="color: white">Team<!--[if gt IE 6]><!--></a><!--<![endif]--> | ||
| + | <!--[if lt IE 7]><table border="0" cellpadding="0" cellspacing="0"><tr><td><![endif]--> | ||
| + | <ul class="DropDownMenu" id="MB1-DDM6"> | ||
| + | <li><a href="https://2008.igem.org/Team:Heidelberg/Team"><span><span>Overview</span></span></a></li> | ||
| + | <li><a href="https://2008.igem.org/Team:Heidelberg/Team_Members#Advisors"><span><span>Advisors</span></span></a></li> | ||
| + | <li><a href="https://2008.igem.org/Team:Heidelberg/Team_Members#Undergraduates"><span><span>Undergraduates</span></span></a></li> | ||
| + | <li><a href="https://2008.igem.org/Team:Heidelberg/Team#There_are_many_Universities_-_and_there_is_Heidelberg_..."><span><span>University</span></span></a></li> | ||
| + | <li><a href="https://2008.igem.org/Team:Heidelberg/Team#DKFZ"><span><span>DKFZ</span></span></a></li> | ||
| + | <li><a href="https://2008.igem.org/Team:Heidelberg/Team#Bioquant"><span><span>BioQuant</span></span></a></li> | ||
| + | <li><a href="https://2008.igem.org/Team:Heidelberg/BioRegioRN"><span><span>BioRegion Rhein-Neckar</span></span></a></li> | ||
| + | </ul> | ||
| + | <!--[if lte IE 6]></td></tr></table></a><![endif]--> | ||
| + | </li> | ||
| + | <li> | ||
| + | <a href="https://2008.igem.org/Team:Heidelberg/Project" style="color: white">Project<!--[if gt IE 6]><!--></a><!--<![endif]--> | ||
| + | <!--[if lt IE 7]><table border="0" cellpadding="0" cellspacing="0"><tr><td><![endif]--> | ||
| + | <ul class="DropDownMenu" id="MB1-DDM1"> | ||
| + | <li><a href="https://2008.igem.org/Team:Heidelberg/Project"><span><span>Overall Project</span></span></a></li> | ||
| + | <li><a href="https://2008.igem.org/Team:Heidelberg/Notebook/material"><span><span>Material & Methods</span></span></a> | ||
| + | <li><a href="https://2008.igem.org/Team:Heidelberg/Project/Sensing"><span><span>Sensing</span></span></a></li> | ||
| + | <li><a href="https://2008.igem.org/Team:Heidelberg/Project/Killing_I"><span><span>Killing I - Phages</span></span></a></li> | ||
| + | <li><a href="https://2008.igem.org/Team:Heidelberg/Project/Killing_II"><span><span>Killing II - Colicin</span></span></a></li> | ||
| + | <li><a href="https://2008.igem.org/Team:Heidelberg/Project/Visualization"><span><span>Visualization</span></span></a></li> | ||
| + | </ul> | ||
| + | <!--[if lte IE 6]></td></tr></table></a><![endif]--> | ||
| + | </li> | ||
| + | |||
| + | <li> | ||
| + | <a href="https://2008.igem.org/Team:Heidelberg/Parts" style="color: white">Parts<!--[if gt IE 6]><!--></a><!--<![endif]--> | ||
| + | <!--[if lt IE 7]><table border="0" cellpadding="0" cellspacing="0"><tr><td><![endif]--> | ||
| + | <ul class="DropDownMenu" id="MB1-DDM2"> | ||
| + | <li><a href="https://2008.igem.org/Team:Heidelberg/Parts"><span><span>Submitted Parts</span></span></a></li> | ||
| + | <li><a href="https://2008.igem.org/Team:Heidelberg/Parts/Characterization"><span><span>Characterization</span></span></a></li> | ||
| + | </ul> | ||
| + | <!--[if lte IE 6]></td></tr></table></a><![endif]--> | ||
| + | </li> | ||
| + | <li> | ||
| + | <a href="https://2008.igem.org/Team:Heidelberg/Modeling" style="color: white">Modeling<!--[if gt IE 6]><!--></a><!--<![endif]--> | ||
| + | <!--[if lt IE 7]><table border="0" cellpadding="0" cellspacing="0"><tr><td><![endif]--> | ||
| + | <ul class="DropDownMenu" id="MB1-DDM3"> | ||
| + | <li><a href="https://2008.igem.org/Team:Heidelberg/Modeling"><span><span>Overview</span></span></a></li> | ||
| + | <li><a href="https://2008.igem.org/Team:Heidelberg/Modeling/Chemotaxis"><span><span>Chemotaxis-Colicin</span></span></a></li> | ||
| + | <li><a href="https://2008.igem.org/Team:Heidelberg/Modeling/Phage"><span><span>Phage Dynamics model</span></span></a></li> | ||
| + | </ul> | ||
| + | <!--[if lte IE 6]></td></tr></table></a><![endif]--> | ||
| + | </li> | ||
| + | <li> | ||
| + | <a href="https://2008.igem.org/Team:Heidelberg/Notebook/Overview" style="color: white">Notebook<!--[if gt IE 6]><!--></a><!--<![endif]--> | ||
| + | <!--[if lt IE 7]><table border="0" cellpadding="0" cellspacing="0"><tr><td><![endif]--> | ||
| + | <ul class="DropDownMenu" id="MB1-DDM5"> | ||
| + | <li><a href="https://2008.igem.org/Team:Heidelberg/Notebook/Sensing_Group/Notebook"><span><span>Sensing ></span></span><!--[if gt IE 6]><!--></a><!--<![endif]--> | ||
| + | <!--[if lt IE 7]><table border="0" cellpadding="0" cellspacing="0"><tr><td><![endif]--> | ||
| + | <ul class="SideMenu" id="MB1-DDM2-SM1"> | ||
| + | <li><a href="https://2008.igem.org/Team:Heidelberg/Notebook/Sensing_Group/Cloning"><span>Cloning strategy</span></a></li> | ||
| + | <li><a href="https://2008.igem.org/Team:Heidelberg/Notebook/Sensing_Group/Notebook"><span>Notebook</span></a></li> | ||
| + | </ul> | ||
| + | <!--[if lte IE 6]></td></tr></table></a><![endif]--> | ||
| + | </li> | ||
| + | |||
| + | <li><a href="https://2008.igem.org/Team:Heidelberg/Notebook/Killing_I/Notebook"><span><span>Killing I - Phages ></span></span><!--[if gt IE 6]><!--></a><!--<![endif]--> | ||
| + | <!--[if lt IE 7]><table border="0" cellpadding="0" cellspacing="0"><tr><td><![endif]--> | ||
| + | <ul class="SideMenu" id="MB1-DDM2-SM2"> | ||
| + | <li><a href="https://2008.igem.org/Team:Heidelberg/Notebook/Killing_I/Cloning"><span>Cloning strategy</span></a></li> | ||
| + | <li><a href="https://2008.igem.org/Team:Heidelberg/Notebook/Killing_I/Notebook"><span>Notebook</span></a></li> | ||
| + | </ul> | ||
| + | <!--[if lte IE 6]></td></tr></table></a><![endif]--> | ||
| + | </li> | ||
| + | <li><a href="https://2008.igem.org/Team:Heidelberg/Notebook/Killing_II/Notebook"><span><span>Killing II - Colicin ></span></span><!--[if gt IE 6]><!--></a><!--<![endif]--> | ||
| + | <!--[if lt IE 7]><table border="0" cellpadding="0" cellspacing="0"><tr><td><![endif]--> | ||
| + | <ul class="SideMenu" id="MB1-DDM2-SM3"> | ||
| + | <li><a href="https://2008.igem.org/Team:Heidelberg/Notebook/Killing_II/Cloning"><span>Cloning strategy</span></a></li> | ||
| + | <li><a href="https://2008.igem.org/Team:Heidelberg/Notebook/Killing_II/Notebook"><span>Notebook</span></a></li> | ||
| + | </ul> | ||
| + | <!--[if lte IE 6]></td></tr></table></a><![endif]--> | ||
| + | </li> | ||
| + | <li><a href="https://2008.igem.org/Team:Heidelberg/Notebook/visualization"><span><span>Visualization</span></span></a> | ||
| + | <li><a href="https://2008.igem.org/Team:Heidelberg/Notebook/material"><span><span>Material & Methods</span></span></a> | ||
| + | <li><a href="https://2008.igem.org/Team:Heidelberg/Notebook/team_meetings"><span><span>Team Meetings</span></span></a> | ||
| + | <li><a href="https://2008.igem.org/Team:Heidelberg/Notebook/seminar"><span><span>Seminar on Synthetic Biology</span></span></a> | ||
| + | </ul> | ||
| + | <!--[if lte IE 6]></td></tr></table></a><![endif]--> | ||
| + | </li> | ||
| + | <li style="width: 160px"> | ||
| + | <a href="https://2008.igem.org/Team:Heidelberg/Human_Practice/Project_Overview" style="color: white">Human Practice<!--[if gt IE 6]><!--></a><!--<![endif]--> | ||
| + | <!--[if lt IE 7]><table border="0" cellpadding="0" cellspacing="0"><tr><td><![endif]--> | ||
| + | <ul class="DropDownMenu" id="MB1-DDM4"> | ||
| + | <li><a href="https://2008.igem.org/Team:Heidelberg/Human_Practice/Project_Overview"><span><span>Project Overview</span></span></a></li> | ||
| + | <li><a href="https://2008.igem.org/Team:Heidelberg/Human_Practice/Phips_the_Phage"><span><span>Phips the Phage</span></span></a></li> | ||
| + | <li><a href="https://2008.igem.org/Team:Heidelberg/Human_Practice/Essay"><span><span>Essay</span></span></a></li> | ||
| + | <li><a href="https://2008.igem.org/Team:Heidelberg/Human_Practice/Surveys"><span><span>Surveys</span></span></a></li> | ||
| + | <li><a href="https://2008.igem.org/Team:Heidelberg/Human_Practice/Open_Day"><span><span>Open Day</span></span></a></li> | ||
| + | <li><a href="https://2008.igem.org/Team:Heidelberg/Human_Practice/Nobel_Prize"><span><span>Nobel Prize</span></span></a></li> | ||
| + | |||
| + | </ul> | ||
| + | <!--[if lte IE 6]></td></tr></table></a><![endif]--> | ||
| + | </li> | ||
| + | |||
| + | <li> | ||
| + | <a href="https://2008.igem.org/Team:Heidelberg/Sponsors" style="color: white">Sponsors</a> | ||
| + | </li> | ||
| + | </ul> | ||
| + | |||
| + | </div> | ||
| + | <div id="header_bottom"><img src="https://static.igem.org/mediawiki/2008/e/ef/Navi_bg.gif" alt="" / ></div> | ||
| + | </body> | ||
| + | </html> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | ==Background== | ||
| + | Interaction of bacterial populations is a challenging and difficult task for modelers. Bacterial strains respond to external chemicals, secrete their own signals for communication, and even produce killing factors for other bacterial species. Many of these processes are non-linear and proceed at different time and space scales. <br> | ||
| + | [[https://2008.igem.org/Team:Heidelberg/Modeling back]] | ||
| + | |||
| + | ==Motivation== | ||
| + | In our project we construct a system of two bacterial strains, in which one population (killer strain) chemotactically senses the chemical signal AI-2 emitted by another population (prey). The killer strain is able to swim towards the prey population using the gradient of AI-2. In the vicinity of prey, the killer strain eliminates the prey. We considered two alternative killing mechanisms: colicin secretion and lambda-phage infection. <br> | ||
| + | [[https://2008.igem.org/Team:Heidelberg/Modeling back]] | ||
| + | |||
| + | ==Our achievements== | ||
| + | We studied the system behavior in space and time using systems of non-linear Partial Differential Equations (PDEs). Systems of PDEs are widely used in science in a variety of problems, from tornadoes prediction to quantum mechanics. | ||
| + | Unlike ordinary differential equations (ODEs), PDEs enable simulation of spatial effects in time, taking into account diffusion effects and spatial fluxes. This is particularly important when the system is not homogeneous. In our case, | ||
| + | the killer population swims towards the prey population, and chemical signals require a certain time to diffuse through the medium. We designed a minimal system of four non-linear reaction-diffusion PDEs to simulate the chemotactic motility of the killer population towards the prey, and the colicin killing process (Fig. 1 (a), (c)). To simulate the system, we approximated the original system of four PDEs by a system of 10,000 ODEs (method of lines), and solved it numerically using custom-written Matlab code. In a more comprehensive PDE model, we added the fifth PDE to reflect the dynamics of AI-1 (Fig. 1 (b), (d)). AI-1 is secreted by the prey cells and adsorbed by the killer cells, in which it activates the production of the colicin and lysis proteins. The killer cells induced by AI-1 lyse and release the colicin into the surrounding media, resulting in the prey elimination. The extended model allowed us to reflect every key component of the system, and to simulate the interaction of the killer and prey populations at different initial spatial distribution and various system parameters. [[https://2008.igem.org/Team:Heidelberg/Modeling back]]<br> <br> | ||
| + | |||
| + | [[Image:Colmod.jpg|center|thumb|733px|'''Figure 1''': The spatial models for chemotaxis of the killer strain to the prey and the colicin-induced killing process. <br> '''(a)''' The minimal model with four components, where AI-1 is assumed to have the same production and diffusion rates as AI-2, both described by equation four.<br>'''(b)'''The corresponding system of four partial | ||
| + | differential equations.<br>'''(c)''' The extended model with five components, where AI-1 dynamics is modeled explicitly.<br>'''(d)'''The corresponding system of five partial differential equations.]] <br> <br> | ||
| + | |||
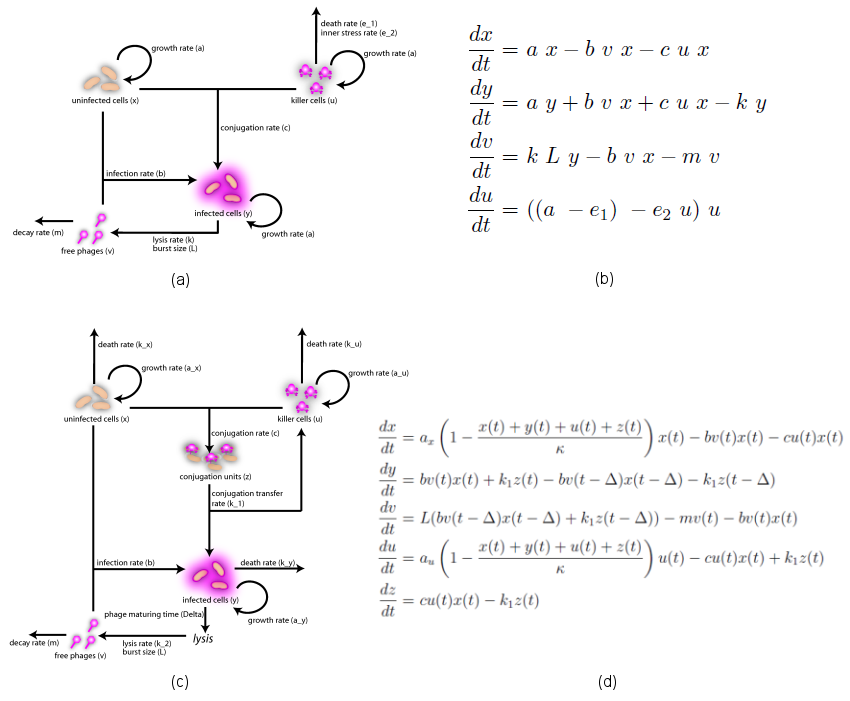
| + | In the case when both populations are well mixed in spatially homogeneous conditions, we modeled another possible killing mechanism – the infection by lambda-phages. Here the prey cells can become infected either by conjugation | ||
| + | with killer cells or by free phages, which in both cases leads to lysis of the prey cells and to a release of multiple new phages, initiating a snowball effect. The assumption of spatial homogeneity allowed us to better resolve the temporal behavior of this complex system. To model the lambda-phage infection, we used a system of delay differential equations (DDEs) (Fig. 2 (a), (c)). Unlike ordinary differential equations (ODEs), DDEs allow the temporal change in concentration to depend on concentrations at earlier time points. In our system, we used this method to reflect the fact that phage maturing cannot be arbitrary fast, but takes a certain time to develop. A careful model verification enabled us to simulate the infection dynamics in detail, including the formation of conjugation complexes and the snowball killing effect. To perform a detailed mathematical analysis, we reduced the full model to a simpler ODE version (Fig. 2 (b), (d)). For the ODE system, we determined its steady states with the linear stability | ||
| + | analysis and investigated their stability with the initial values analysis, enabling to predict different system regimes upon given parameters and initial conditions. [[https://2008.igem.org/Team:Heidelberg/Modeling back]]<br> <br> <br> | ||
| + | |||
| + | [[Image:PhageSummary.png|center|thumb|733px|'''Figure 2''': The models of ''E. Coli'' infection by lambda-phages.<br/> '''(a)''' Scheme of the simplified model. <br /> '''(b)''' The ODE system which describes the simplified model of lambda-phage infection. <br /> '''(c)''' Scheme of the full model. <br /> '''(d)''' The DDE system which describes the full model of the phage infection.]] | ||
| + | |||
| + | ==Modeling Results== | ||
| + | If you are interested in more details concerning <br><br> | ||
| + | our modeling part of '''Spatially Resolved Modeling of Colicin-Induced Killing and Chemotactic Sensing in ''E. coli''''', please click [https://2008.igem.org/Team:Heidelberg/Modeling/Chemotaxis here] <br><br> | ||
| + | our modeling part of '''Phage-induced infection dynamics of ''E. coli'' ''', please click [[Media:Phagedocu.pdf|here]] <br><br> | ||
| + | '''Status reports''' of our group, please click [https://2008.igem.org/Team:Heidelberg/Notebook/team_meetings#Team_Meetings here].<br><br> | ||
| + | To download this '''summary''' of our modeling project, please click [[Media:Summary.pdf|here]] | ||
| + | |||
| + | [[https://2008.igem.org/Team:Heidelberg/Modeling back]] | ||
Latest revision as of 23:06, 29 October 2008


Background
Interaction of bacterial populations is a challenging and difficult task for modelers. Bacterial strains respond to external chemicals, secrete their own signals for communication, and even produce killing factors for other bacterial species. Many of these processes are non-linear and proceed at different time and space scales.
[back]
Motivation
In our project we construct a system of two bacterial strains, in which one population (killer strain) chemotactically senses the chemical signal AI-2 emitted by another population (prey). The killer strain is able to swim towards the prey population using the gradient of AI-2. In the vicinity of prey, the killer strain eliminates the prey. We considered two alternative killing mechanisms: colicin secretion and lambda-phage infection.
[back]
Our achievements
We studied the system behavior in space and time using systems of non-linear Partial Differential Equations (PDEs). Systems of PDEs are widely used in science in a variety of problems, from tornadoes prediction to quantum mechanics.
Unlike ordinary differential equations (ODEs), PDEs enable simulation of spatial effects in time, taking into account diffusion effects and spatial fluxes. This is particularly important when the system is not homogeneous. In our case,
the killer population swims towards the prey population, and chemical signals require a certain time to diffuse through the medium. We designed a minimal system of four non-linear reaction-diffusion PDEs to simulate the chemotactic motility of the killer population towards the prey, and the colicin killing process (Fig. 1 (a), (c)). To simulate the system, we approximated the original system of four PDEs by a system of 10,000 ODEs (method of lines), and solved it numerically using custom-written Matlab code. In a more comprehensive PDE model, we added the fifth PDE to reflect the dynamics of AI-1 (Fig. 1 (b), (d)). AI-1 is secreted by the prey cells and adsorbed by the killer cells, in which it activates the production of the colicin and lysis proteins. The killer cells induced by AI-1 lyse and release the colicin into the surrounding media, resulting in the prey elimination. The extended model allowed us to reflect every key component of the system, and to simulate the interaction of the killer and prey populations at different initial spatial distribution and various system parameters. [back]
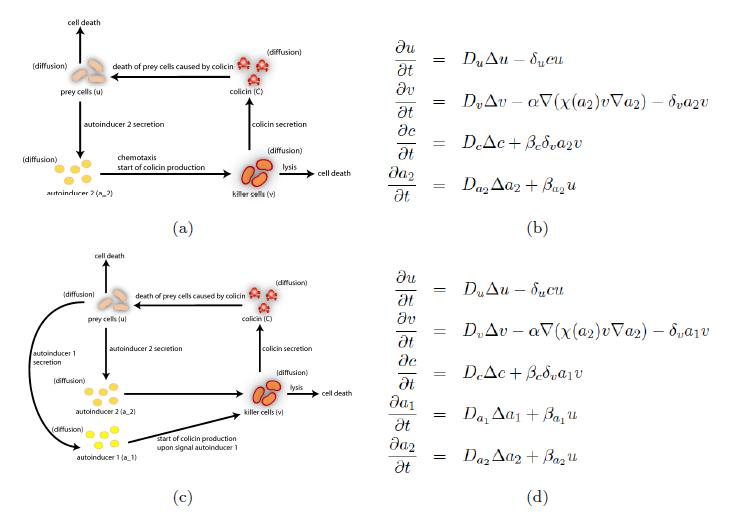
(a) The minimal model with four components, where AI-1 is assumed to have the same production and diffusion rates as AI-2, both described by equation four.
(b)The corresponding system of four partial differential equations.
(c) The extended model with five components, where AI-1 dynamics is modeled explicitly.
(d)The corresponding system of five partial differential equations.
In the case when both populations are well mixed in spatially homogeneous conditions, we modeled another possible killing mechanism – the infection by lambda-phages. Here the prey cells can become infected either by conjugation
with killer cells or by free phages, which in both cases leads to lysis of the prey cells and to a release of multiple new phages, initiating a snowball effect. The assumption of spatial homogeneity allowed us to better resolve the temporal behavior of this complex system. To model the lambda-phage infection, we used a system of delay differential equations (DDEs) (Fig. 2 (a), (c)). Unlike ordinary differential equations (ODEs), DDEs allow the temporal change in concentration to depend on concentrations at earlier time points. In our system, we used this method to reflect the fact that phage maturing cannot be arbitrary fast, but takes a certain time to develop. A careful model verification enabled us to simulate the infection dynamics in detail, including the formation of conjugation complexes and the snowball killing effect. To perform a detailed mathematical analysis, we reduced the full model to a simpler ODE version (Fig. 2 (b), (d)). For the ODE system, we determined its steady states with the linear stability
analysis and investigated their stability with the initial values analysis, enabling to predict different system regimes upon given parameters and initial conditions. [back]
Modeling Results
If you are interested in more details concerning
our modeling part of Spatially Resolved Modeling of Colicin-Induced Killing and Chemotactic Sensing in E. coli, please click here
our modeling part of Phage-induced infection dynamics of E. coli , please click here
Status reports of our group, please click here.
To download this summary of our modeling project, please click here
[back]
 "
"