TUDelft/25 September 2008
From 2008.igem.org
(New page: {{Template:TUDelftiGEM2008}} {{Template:TUDelftiGEM2008_menu_home}} {{Template:TUDelftiGEM2008_calendar}} =September 25th 2008== ==Luciferase Assay== ===Sample preparation=== All the cul...) |
(→Luciferase Assay) |
||
| (8 intermediate revisions not shown) | |||
| Line 3: | Line 3: | ||
{{Template:TUDelftiGEM2008_calendar}} | {{Template:TUDelftiGEM2008_calendar}} | ||
| - | =September 25th 2008 | + | =September 25th 2008= |
==Luciferase Assay== | ==Luciferase Assay== | ||
===Sample preparation=== | ===Sample preparation=== | ||
| - | All the cultures growing had reached an OD of about 0.4 in the morning. When 3 hours later the OD's had stayed practically constant, we lysed them. Also the cultures of K115036 growing at | + | All the cultures growing at 42ºC had reached an OD of about 0.4 in the morning. When 3 hours later the OD's had stayed practically constant, we lysed them. Also the cultures of K115036 growing at 30 and 37ºC have been lysed. Also has K115036 been pelleted and frozen at -20ºC for miniprepping later. |
| - | Of all cultures lysed, 10x and 100x dilutions have been made in H2O. 100x dilutions showed to be in the linear range of the BCA protein assay in a preliminary test, so these were used for total protein determinations. | + | Of all cultures lysed, 10x and 100x dilutions have been made in H2O. 100x dilutions showed to be in the linear range of the BCA protein assay in a preliminary test, so these were used for total protein determinations. |
===Results=== | ===Results=== | ||
| - | + | '''Note that these results are not reliable due to interference of the lysis buffer with the total protein content measurements''' | |
| - | + | The strains measured contained one of the following constructs: P1010 in pSB1AT3, K115012, K115029, K115031, K115032 (which had a dubious PCR product, it was lane 32 1' [https://2008.igem.org/Image:TUDelftColonyPCR230908.jpg here]), K115034 and K115036. Only K115036 was not measured at 26ºC. | |
| - | The normalized results of the luciferase assay (corrected for protein content) are displayed in graph 1. | + | Unfortunately the reader had some problems, making it impossible to do good measurements on K115031. Also did we find out pure 100x diluted lysis buffer gives quite high values in the total protein assay, so in next experiments we'll need to correct our calibration curve for this as well. In an extra test of R0080, we grew some non-induced samples at 37ºC (K115034 and K115012), but these also had high expression of luciferase, so it will not be possible to grow and induce at different temperatures with R0080. |
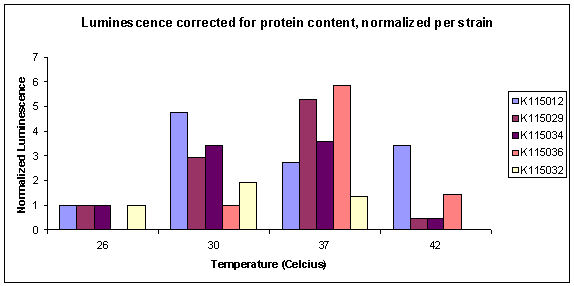
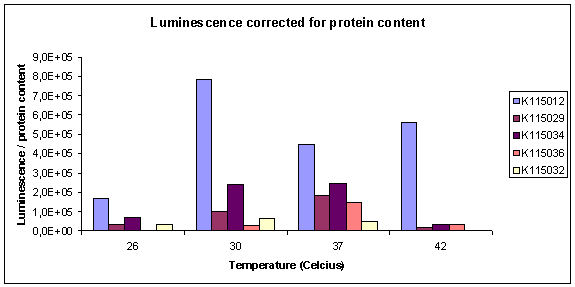
| + | The normalized results of the luciferase assay (corrected for protein content) are displayed in graph 1. The total luminescence corrected by protein content can be seen in graph 2. All strains are normalized for T=26ºC with the exception of K115036, where this datapoint was missing. | ||
| + | *N.B.: Before October 13, we normalized all the datapoints for luminescence of strain K115012 at the same temperature. After some discussion we decided it would be better to normalize luminescence per strain, for lowest available temperature. | ||
| - | [[Image:TUDelft_080925Luciferase.jpg | | + | [[Image:TUDelft_080925Luciferase.jpg | 550px]] |
<i>Graph 1: Normalized Luciferase Activity measurements.</i> | <i>Graph 1: Normalized Luciferase Activity measurements.</i> | ||
| + | It can be seen that relative expression goes down from 37 to 26ºC. This is what we would hope to see. Unfortunately, the datapoints for the reference strain K115012 were not very good. Especially the point for T=37ºC is unexpected. The true corrected datapoints are visible in graph 2. We didn't do the luciferase assay in duplo, therefore data points are also unreliable. At high temperature, K115012 seems to remain a lot more stable than the thermosensitive constructs. More measurements must be done to obtain more datapoints at different temperatures to verify the observed behaviour between 37 and 26ºC. | ||
| + | |||
| + | [[Image:TUDelft_080925Luciferase2.jpg | 550px]] | ||
| + | |||
| + | <i>Graph 2: Luminescence measured, corrected for protein content of the sample.</i> | ||
| + | |||
| + | ==Miniprep K115036== | ||
| + | To obtain a [http://partsregistry.org/wiki/index.php?title=Part:BBa_K115036 K115036] plasmid stock, we've frozen a cell pellet for later miniprepping. | ||
{{Template:TUDelftiGEM2008_sidebar}} | {{Template:TUDelftiGEM2008_sidebar}} | ||
Latest revision as of 15:30, 28 October 2008
| July | ||||||
| M | T | W | T | F | S | S |
| 1 | 2 | 3 | 4 | 5 | 6 | |
| 7 | 8 | 9 | 10 | 11 | 12 | 13 |
| 14 | 15 | 16 | 17 | 18 | 19 | 20 |
| 21 | 22 | 23 | 24 | 25 | 26 | 27 |
| 28 | 29 | 30 | 31 | |||
| August | ||||||
| M | T | W | T | F | S | S |
| 1 | 2 | 3 | ||||
| 4 | 5 | 6 | 7 | 8 | 9 | 10 |
| 11 | 12 | 13 | 14 | 15 | 16 | 17 |
| 18 | 19 | 20 | 21 | 22 | 23 | 24 |
| 25 | 26 | 27 | 28 | 29 | 30 | 31 |
| September | ||||||
| M | T | W | T | F | S | S |
| [http://2008.igem.org/TUDelft/1_September_2008 1] | [http://2008.igem.org/TUDelft/2_September_2008 2] | [http://2008.igem.org/TUDelft/3_September_2008 3] | [http://2008.igem.org/TUDelft/4_September_2008 4] | [http://2008.igem.org/TUDelft/5_September_2008 5] | [http://2008.igem.org/wiki/index.php?title=TUDelft/6_September_2008&action=edit 6] | [http://2008.igem.org/wiki/index.php?title=TUDelft/7_September_2008&action=edit 7] |
| [http://2008.igem.org/TUDelft/8_September_2008 8] | [http://2008.igem.org/TUDelft/9_September_2008 9] | [http://2008.igem.org/TUDelft/10_September_2008 10] | [http://2008.igem.org/TUDelft/11_September_2008 11] | [http://2008.igem.org/TUDelft/12_September_2008 12] | [http://2008.igem.org/wiki/index.php?title=TUDelft/13_September_2008&action=edit 13] | [http://2008.igem.org/wiki/index.php?title=TUDelft/14_September_2008&action=edit 14] |
| [http://2008.igem.org/TUDelft/15_September_2008 15] | [http://2008.igem.org/TUDelft/16_September_2008 16] | [http://2008.igem.org/TUDelft/17_September_2008 17] | [http://2008.igem.org/TUDelft/18_September_2008 18] | [http://2008.igem.org/TUDelft/19_September_2008 19] | [http://2008.igem.org/wiki/index.php?title=TUDelft/20_September_2008&action=edit 20] | [http://2008.igem.org/wiki/index.php?title=TUDelft/21_September_2008&action=edit 21] |
| [http://2008.igem.org/TUDelft/22_September_2008 22] | [http://2008.igem.org/TUDelft/23_September_2008 23] | [http://2008.igem.org/TUDelft/24_September_2008 24] | [http://2008.igem.org/TUDelft/25_September_2008 25] | [http://2008.igem.org/wiki/index.php?title=TUDelft/26_September_2008&action=edit 26] | [http://2008.igem.org/wiki/index.php?title=TUDelft/27_September_2008&action=edit 27] | [http://2008.igem.org/wiki/index.php?title=TUDelft/28_September_2008&action=edit 28] |
| [http://2008.igem.org/TUDelft/29_September_2008 29] | [http://2008.igem.org/TUDelft/30_September_2008 30] | |||||
| October | ||||||
| M | T | W | T | F | S | S |
| [http://2008.igem.org/TUDelft/1_October_2008 1] | [http://2008.igem.org/TUDelft/2_October_2008 2] | [http://2008.igem.org/TUDelft/3_October_2008 3] | [http://2008.igem.org/wiki/index.php?title=TUDelft/4_October_2008&action=edit 4] | [http://2008.igem.org/wiki/index.php?title=TUDelft/5_October_2008&action=edit 5] | ||
| [http://2008.igem.org/TUDelft/6_October_2008 6] | [http://2008.igem.org/TUDelft/7_October_2008 7] | [http://2008.igem.org/TUDelft/8_October_2008 8] | [http://2008.igem.org/TUDelft/9_October_2008 9] | [http://2008.igem.org/TUDelft/10_October_2008 10] | [http://2008.igem.org/wiki/index.php?title=TUDelft/11_October_2008&action=edit 11] | [http://2008.igem.org/wiki/index.php?title=TUDelft/12_October_2008&action=edit 12] |
| [http://2008.igem.org/TUDelft/13_October_2008 13] | [http://2008.igem.org/TUDelft/14_October_2008 14] | [http://2008.igem.org/TUDelft/15_October_2008 15] | [http://2008.igem.org/TUDelft/16_October_2008 16] | [http://2008.igem.org/TUDelft/17_October_2008 17] | [http://2008.igem.org/wiki/index.php?title=TUDelft/18_October_2008&action=edit 18] | [http://2008.igem.org/wiki/index.php?title=TUDelft/19_October_2008&action=edit 19] |
| [http://2008.igem.org/TUDelft/20_October_2008 20] | [http://2008.igem.org/TUDelft/21_October_2008 21] | [http://2008.igem.org/TUDelft/22_October_2008 22] | [http://2008.igem.org/TUDelft/23_October_2008 23] | [http://2008.igem.org/TUDelft/24_October_2008 24] | [http://2008.igem.org/wiki/index.php?title=TUDelft/25_October_2008&action=edit 25] | [http://2008.igem.org/wiki/index.php?title=TUDelft/26_October_2008&action=edit 26] |
| [http://2008.igem.org/wiki/index.php?title=TUDelft/27_October_2008&action=edit 27] | [http://2008.igem.org/wiki/index.php?title=TUDelft/28_October_2008&action=edit 28] | [http://2008.igem.org/wiki/index.php?title=TUDelft/29_October_2008&action=edit 29] | [http://2008.igem.org/wiki/index.php?title=TUDelft/30_October_2008&action=edit 30] | [http://2008.igem.org/wiki/index.php?title=TUDelft/31_October_2008&action=edit 31] | ||
Contents |
September 25th 2008
Luciferase Assay
Sample preparation
All the cultures growing at 42ºC had reached an OD of about 0.4 in the morning. When 3 hours later the OD's had stayed practically constant, we lysed them. Also the cultures of K115036 growing at 30 and 37ºC have been lysed. Also has K115036 been pelleted and frozen at -20ºC for miniprepping later.
Of all cultures lysed, 10x and 100x dilutions have been made in H2O. 100x dilutions showed to be in the linear range of the BCA protein assay in a preliminary test, so these were used for total protein determinations.
Results
Note that these results are not reliable due to interference of the lysis buffer with the total protein content measurements
The strains measured contained one of the following constructs: P1010 in pSB1AT3, K115012, K115029, K115031, K115032 (which had a dubious PCR product, it was lane 32 1' here), K115034 and K115036. Only K115036 was not measured at 26ºC.
Unfortunately the reader had some problems, making it impossible to do good measurements on K115031. Also did we find out pure 100x diluted lysis buffer gives quite high values in the total protein assay, so in next experiments we'll need to correct our calibration curve for this as well. In an extra test of R0080, we grew some non-induced samples at 37ºC (K115034 and K115012), but these also had high expression of luciferase, so it will not be possible to grow and induce at different temperatures with R0080. The normalized results of the luciferase assay (corrected for protein content) are displayed in graph 1. The total luminescence corrected by protein content can be seen in graph 2. All strains are normalized for T=26ºC with the exception of K115036, where this datapoint was missing.
- N.B.: Before October 13, we normalized all the datapoints for luminescence of strain K115012 at the same temperature. After some discussion we decided it would be better to normalize luminescence per strain, for lowest available temperature.
Graph 1: Normalized Luciferase Activity measurements.
It can be seen that relative expression goes down from 37 to 26ºC. This is what we would hope to see. Unfortunately, the datapoints for the reference strain K115012 were not very good. Especially the point for T=37ºC is unexpected. The true corrected datapoints are visible in graph 2. We didn't do the luciferase assay in duplo, therefore data points are also unreliable. At high temperature, K115012 seems to remain a lot more stable than the thermosensitive constructs. More measurements must be done to obtain more datapoints at different temperatures to verify the observed behaviour between 37 and 26ºC.
Graph 2: Luminescence measured, corrected for protein content of the sample.
Miniprep K115036
To obtain a [http://partsregistry.org/wiki/index.php?title=Part:BBa_K115036 K115036] plasmid stock, we've frozen a cell pellet for later miniprepping.
 "
"