Mutation Model
From 2008.igem.org
(Difference between revisions)
(→Part I Mutation Model) |
|||
| Line 1: | Line 1: | ||
=Part I Mutation Model= | =Part I Mutation Model= | ||
==Hypothesis== | ==Hypothesis== | ||
| - | #The mutation efficiency should remain a continuous curve along reproduction, or along time. Here we discrete the mutation rate to be linear function according to the replication time (), which in fact create several discontiguous point. | + | #The mutation efficiency from the initial gene to the target gene should remain a continuous curve along reproduction, or along time. Here we discrete the mutation rate to be linear function according to the replication time (), which in fact create several discontiguous point. |
#DNA lesion repair rate appears high enough in replication than other cell activities, so that the DNA lesion repair rate can be conformed to background mutations except in replication process. | #DNA lesion repair rate appears high enough in replication than other cell activities, so that the DNA lesion repair rate can be conformed to background mutations except in replication process. | ||
#The whole yeast is large enough to obey the statistics rules | #The whole yeast is large enough to obey the statistics rules | ||
| Line 7: | Line 7: | ||
#Probability Model <br>For each DNA sequence status, we take down a probability matrix for the coding sequence, which gives out each base appearance possibility on every site. The structure on each site is 5*1 matrix,(P<sub>a</sub>,P<sub>t</sub>,P<sub>c</sub>,P<sub>g</sub>,P<sub>u</sub>), and ΣP=1. Therefore, the whole sequence status is recorded as 5*length matrix.E.g. For the Gal4 DNA, with a length of 2646, one status is a 5*2646 matrix. | #Probability Model <br>For each DNA sequence status, we take down a probability matrix for the coding sequence, which gives out each base appearance possibility on every site. The structure on each site is 5*1 matrix,(P<sub>a</sub>,P<sub>t</sub>,P<sub>c</sub>,P<sub>g</sub>,P<sub>u</sub>), and ΣP=1. Therefore, the whole sequence status is recorded as 5*length matrix.E.g. For the Gal4 DNA, with a length of 2646, one status is a 5*2646 matrix. | ||
#Scanning Model<br>The hotbox appearance rate on a specific site i is calculated as below. | #Scanning Model<br>The hotbox appearance rate on a specific site i is calculated as below. | ||
| + | {|align="left" | ||
| + | |[[Image:Peking hotbox search.jpg]] | ||
| + | |}<br><br><br> | ||
| + | W: a, t; R: a, g; H: a, c, g;<br> | ||
| + | Hotbox appearrance rate P<sub>H</sub>=P<sub>W</sub><sup>(i-2)</sup>P<sub>R</sub><sup>(i-2)</sup>P<sub>C</sub><sup>(i-2)</sup>P<sub>H</sub><sup>(i-2)</sup>, and here (i) refers to the site i. | ||
| + | #Model on DNA lesion repair | ||
| + | As reviewed by the paper, the U:G mismatch DNA lesion will be repaired in three pathways. | ||
| + | {|align="centre" | ||
| + | |[[Image:Peking Repair.jpg]] | ||
| + | |} | ||
| + | {|align="right" | ||
| + | |''VH Odegard, DG Schatz. Targeting of somatic hypermutation. Nat Rev Immunol. 2006 Aug;6(8):573-83'' | ||
| + | |} | ||
Revision as of 17:35, 29 October 2008
Part I Mutation Model
Hypothesis
- The mutation efficiency from the initial gene to the target gene should remain a continuous curve along reproduction, or along time. Here we discrete the mutation rate to be linear function according to the replication time (), which in fact create several discontiguous point.
- DNA lesion repair rate appears high enough in replication than other cell activities, so that the DNA lesion repair rate can be conformed to background mutations except in replication process.
- The whole yeast is large enough to obey the statistics rules
Model Construction
- Probability Model
For each DNA sequence status, we take down a probability matrix for the coding sequence, which gives out each base appearance possibility on every site. The structure on each site is 5*1 matrix,(Pa,Pt,Pc,Pg,Pu), and ΣP=1. Therefore, the whole sequence status is recorded as 5*length matrix.E.g. For the Gal4 DNA, with a length of 2646, one status is a 5*2646 matrix. - Scanning Model
The hotbox appearance rate on a specific site i is calculated as below.

|
W: a, t; R: a, g; H: a, c, g;
Hotbox appearrance rate PH=PW(i-2)PR(i-2)PC(i-2)PH(i-2), and here (i) refers to the site i.
- Model on DNA lesion repair
As reviewed by the paper, the U:G mismatch DNA lesion will be repaired in three pathways.
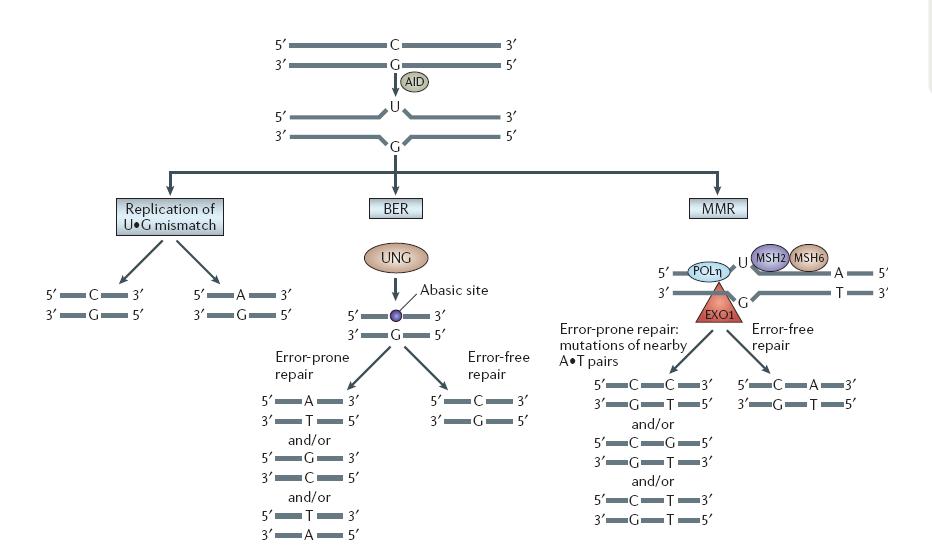
|
| VH Odegard, DG Schatz. Targeting of somatic hypermutation. Nat Rev Immunol. 2006 Aug;6(8):573-83 |
 "
"