Newcastle University Drylab/4 June 2008
From 2008.igem.org
(New page: {{:Team:Newcastle_University/DrylabCalendarHeader}} == 4 June 2008 == ===Mark=== Spent the majority of the day hassling with the EA, and getting the bugs out of the program. Got the bugs...) |
|||
| Line 6: | Line 6: | ||
Spent the majority of the day hassling with the EA, and getting the bugs out of the program. Got the bugs out of the system and the basic EA now works and evolves the hidden nodes until the threshold value is met. | Spent the majority of the day hassling with the EA, and getting the bugs out of the program. Got the bugs out of the system and the basic EA now works and evolves the hidden nodes until the threshold value is met. | ||
| - | Code sample for the mutate method is shown below: public void mutate() | + | Code sample for the mutate method is shown below: |
| + | <pre>public void mutate() | ||
{ | { | ||
| Line 26: | Line 27: | ||
//System.out.println(promoter); | //System.out.println(promoter); | ||
promoter.setStrength(value); } | promoter.setStrength(value); } | ||
| - | }Spent some time with Matt and Morgan doing user stories for the interface. Wrote some down for the EA itself. | + | }</pre> |
| + | |||
| + | Spent some time with Matt and Morgan doing user stories for the interface. Wrote some down for the EA itself. | ||
| Line 60: | Line 63: | ||
===Morgan=== | ===Morgan=== | ||
| - | Edited the | + | Edited the [http://bioinf.ncl.ac.uk/iGEM/wiki/doku.php?id=java_tutorial_and_exercises#constraints_repository ConstraintsRepository exercise] in the Java exercises to reflect more of what we discussed yesterday. |
Back to the CellML Tutorial today. Part 2! Now we're adding components and interactions. | Back to the CellML Tutorial today. Part 2! Now we're adding components and interactions. | ||
| Line 66: | Line 69: | ||
Constants are indicated in equations in COR just by using numbers, followed by the units in curly brackets. | Constants are indicated in equations in COR just by using numbers, followed by the units in curly brackets. | ||
| - | < | + | <pre> |
V = 1{millivolt}/(1{dimensionless}+exp((C+1.6{uM})/-1.42{uM})); | V = 1{millivolt}/(1{dimensionless}+exp((C+1.6{uM})/-1.42{uM})); | ||
| - | </ | + | </pre> |
In the cellml, the <cn> tag is used to indicate constants, along with their units. | In the cellml, the <cn> tag is used to indicate constants, along with their units. | ||
| - | < | + | <pre> |
<cn cellml:units="millivolt">1</cn> | <cn cellml:units="millivolt">1</cn> | ||
| - | </ | + | </pre> |
Incidently, the <ci> tag is used for constants in the MathML equations. | Incidently, the <ci> tag is used for constants in the MathML equations. | ||
| - | Mapping is used to connect two components together. The relevant variables have to be connected to one another using the map components syntax. The varibles must include the phrase | + | Mapping is used to connect two components together. The relevant variables have to be connected to one another using the map components syntax. The varibles must include the phrase ''pub: in'' or ''pub: out'' depending on whether they're accepting values or donating values. The ''pub: in'' variable also should be initialized, I think. Oh, and they have to be in the right order. Map between variables outVar and inVar. It doesn't work the other way around. |
| - | < | + | <pre> |
def map between firstComponent and secondComponent for | def map between firstComponent and secondComponent for | ||
vars outVar and inVar; | vars outVar and inVar; | ||
enddef; | enddef; | ||
| - | </ | + | </pre> |
and the equivalent CellML is: | and the equivalent CellML is: | ||
| - | < | + | <pre> |
<connection xmlns="http://www.cellml.org/cellml/1.0#"> | <connection xmlns="http://www.cellml.org/cellml/1.0#"> | ||
<map_components component_1="firstComponent" component_2="secondComponent"/> | <map_components component_1="firstComponent" component_2="secondComponent"/> | ||
<map_variables variable_1="outVar" variable_2="inVar"/> | <map_variables variable_1="outVar" variable_2="inVar"/> | ||
</connection> | </connection> | ||
| - | </ | + | </pre> |
| - | A'ight, that works. Now Tutorial 3. Since COR doesn't handle CellML 1.1, I had to edit it in WordPad and then run it in | + | A'ight, that works. Now Tutorial 3. Since COR doesn't handle CellML 1.1, I had to edit it in WordPad and then run it in [http://www.cellml.org/tools/pcenv/ PCEnv]. Mike and James installed it on my laptop when they were here, so luckily I only had to do a search and there it was. |
| - | It appears that you can import both CellML 1.0 and 1.1, as long as your 'main' file is of 1.1 type. COR refuses to let you save something that's mapped to | + | It appears that you can import both CellML 1.0 and 1.1, as long as your 'main' file is of 1.1 type. COR refuses to let you save something that's mapped to ''pub: out'' or ''pub: in'' without some kind of internal mapping, though, so I guess you have to manually edit all the files anyway. |
The linking between the files is done with XLinks. I guess it was kind of inevitable that I'd run into them again. Maybe I'll actually learn them this time. In order to change over a 1.0 file, you have to change all the model namespaces (except metadata) to 1.1, and then add a new one: xmlns:xlink="http://www.w3.org/1999/xlink. Then you use the XLink stuff to import the other file, like so: | The linking between the files is done with XLinks. I guess it was kind of inevitable that I'd run into them again. Maybe I'll actually learn them this time. In order to change over a 1.0 file, you have to change all the model namespaces (except metadata) to 1.1, and then add a new one: xmlns:xlink="http://www.w3.org/1999/xlink. Then you use the XLink stuff to import the other file, like so: | ||
| - | < | + | <pre> |
<import xlink:href="filenameOfModelToBeImported.cellml"> | <import xlink:href="filenameOfModelToBeImported.cellml"> | ||
<component name="intracellular" component_ref="firstComponent" /> | <component name="intracellular" component_ref="firstComponent" /> | ||
</import> | </import> | ||
| - | </ | + | </pre> |
I suppose for each component that you import, you need to call it something different. Then in the mapping you can just refer to it like any other component. | I suppose for each component that you import, you need to call it something different. Then in the mapping you can just refer to it like any other component. | ||
| - | < | + | <pre> |
<connection xmlns="http://www.cellml.org/cellml/1.0#"> | <connection xmlns="http://www.cellml.org/cellml/1.0#"> | ||
<map_components component_1="intracellular" component_2="secondComponent"/> | <map_components component_1="intracellular" component_2="secondComponent"/> | ||
<map_variables variable_1="outVar" variable_2="inVar"/> | <map_variables variable_1="outVar" variable_2="inVar"/> | ||
</connection> | </connection> | ||
| - | </ | + | </pre> |
Hm, that's odd that the namespace is still 1.0 and it works. OH well. Yay, I got it to run on PCEnv! Whoever designed that program though didn't have much idea how to design user interfaces... | Hm, that's odd that the namespace is still 1.0 and it works. OH well. Yay, I got it to run on PCEnv! Whoever designed that program though didn't have much idea how to design user interfaces... | ||
| Line 122: | Line 125: | ||
Oh yeah, I remember why I couldn't persuade it to work. It only compiles on Linux. Sigh. In my opinion, a program that can only compile in Linux is just as useless as one that can only compile in Windows. But that's probably my Java background talking. | Oh yeah, I remember why I couldn't persuade it to work. It only compiles on Linux. Sigh. In my opinion, a program that can only compile in Linux is just as useless as one that can only compile in Windows. But that's probably my Java background talking. | ||
| - | Alright, back to | + | Alright, back to [http://jdesigner.sourceforge.net/ SBW] then. Hehe, the SBML support module is called NOM. [http://icanhascheezburger.com/2007/11/22/nom-nom-nom-nom/ NOM NOM NOM]. |
I asked Neil for the documents defining promoters and differeent parts, and he gave me several documents to read. | I asked Neil for the documents defining promoters and differeent parts, and he gave me several documents to read. | ||
| - | AGH. | + | AGH. |
| - | < | + | |
| + | <pre> | ||
[Error] ..\..\Users\Morgan\regulatory.cellml: the free variable '' is not defined anywhere | [Error] ..\..\Users\Morgan\regulatory.cellml: the free variable '' is not defined anywhere | ||
| - | </ | + | </pre> |
Why! | Why! | ||
| - | |||
| - | |||
===Nina=== | ===Nina=== | ||
Latest revision as of 12:44, 13 October 2008
Newcastle University
GOLD MEDAL WINNER 2008
| Home | Team | Original Aims | Software | Modelling | Proof of Concept Brick | Wet Lab | Conclusions |
|---|
Home >> Dry Lab >> Dry Lab Journal
|
|
|
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
4 June 2008
Mark
Spent the majority of the day hassling with the EA, and getting the bugs out of the program. Got the bugs out of the system and the basic EA now works and evolves the hidden nodes until the threshold value is met.
Code sample for the mutate method is shown below:
public void mutate()
{
int mutateThreshold = 4;
Random random = new Random();
int mutate =random.nextInt();
int change = 0;
double value = 0; if (mutate<=mutateThreshold)
{ }
else {
for (int i=0; i<hidden.length; i++)
{
value = random.nextInt(5);
change = random.nextInt(5);
Node node = hidden[i];
//System.out.println("Node value" +hidden[i]);
Promoter[] barry = node.getCarry();
//System.out.println("Node getcarry" + node.getCarry()); Promoter promoter = barry[change];
//System.out.println(promoter);
promoter.setStrength(value); }
}
Spent some time with Matt and Morgan doing user stories for the interface. Wrote some down for the EA itself.
Megan
Research into biological concept (S.pneumoniae). It is vital to work out whether or not the vnc system can be used, the only problem continues to be this mystery repressor.... As far as I can see there may be a gene that mediates the overall pathway however within the two-component system I do not think that there is a specific inhibitor.
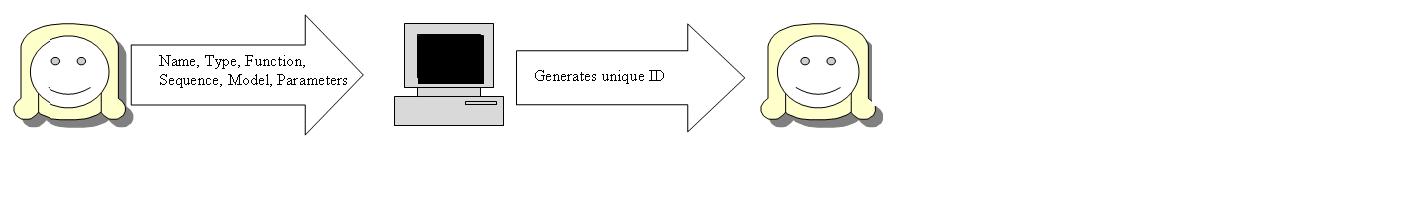
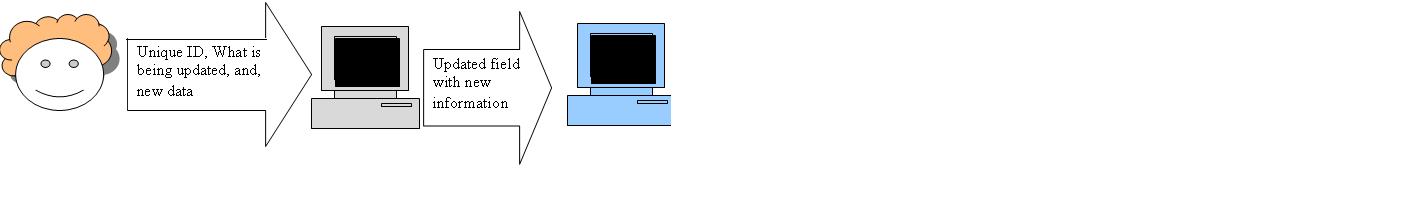
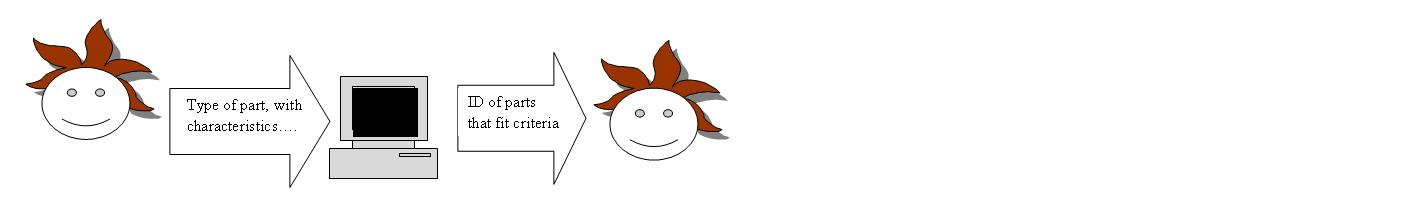
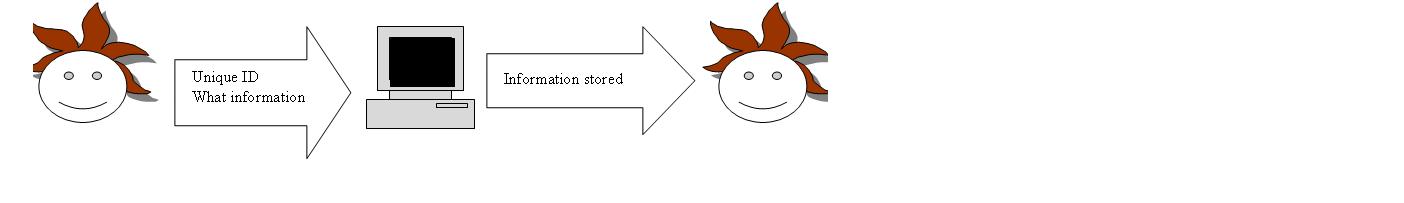
Outline the cases for the 4 user stories from yesterday. These user stories are; New part, Update of existing part, Query/retrieve ID, get Part information(based on ID)
New Part
Update
Retrieve IDs
Resolve ID
These are the four user cases simplified by schematics.
Morgan
Edited the ConstraintsRepository exercise in the Java exercises to reflect more of what we discussed yesterday.
Back to the CellML Tutorial today. Part 2! Now we're adding components and interactions.
Constants are indicated in equations in COR just by using numbers, followed by the units in curly brackets.
V = 1{millivolt}/(1{dimensionless}+exp((C+1.6{uM})/-1.42{uM}));
In the cellml, the <cn> tag is used to indicate constants, along with their units.
<cn cellml:units="millivolt">1</cn>
Incidently, the <ci> tag is used for constants in the MathML equations.
Mapping is used to connect two components together. The relevant variables have to be connected to one another using the map components syntax. The varibles must include the phrase pub: in or pub: out depending on whether they're accepting values or donating values. The pub: in variable also should be initialized, I think. Oh, and they have to be in the right order. Map between variables outVar and inVar. It doesn't work the other way around.
def map between firstComponent and secondComponent for vars outVar and inVar; enddef;
and the equivalent CellML is:
<connection xmlns="http://www.cellml.org/cellml/1.0#">
<map_components component_1="firstComponent" component_2="secondComponent"/>
<map_variables variable_1="outVar" variable_2="inVar"/>
</connection>
A'ight, that works. Now Tutorial 3. Since COR doesn't handle CellML 1.1, I had to edit it in WordPad and then run it in PCEnv. Mike and James installed it on my laptop when they were here, so luckily I only had to do a search and there it was.
It appears that you can import both CellML 1.0 and 1.1, as long as your 'main' file is of 1.1 type. COR refuses to let you save something that's mapped to pub: out or pub: in without some kind of internal mapping, though, so I guess you have to manually edit all the files anyway.
The linking between the files is done with XLinks. I guess it was kind of inevitable that I'd run into them again. Maybe I'll actually learn them this time. In order to change over a 1.0 file, you have to change all the model namespaces (except metadata) to 1.1, and then add a new one: xmlns:xlink="http://www.w3.org/1999/xlink. Then you use the XLink stuff to import the other file, like so:
<import xlink:href="filenameOfModelToBeImported.cellml"> <component name="intracellular" component_ref="firstComponent" /> </import>
I suppose for each component that you import, you need to call it something different. Then in the mapping you can just refer to it like any other component.
<connection xmlns="http://www.cellml.org/cellml/1.0#"> <map_components component_1="intracellular" component_2="secondComponent"/> <map_variables variable_1="outVar" variable_2="inVar"/> </connection>
Hm, that's odd that the namespace is still 1.0 and it works. OH well. Yay, I got it to run on PCEnv! Whoever designed that program though didn't have much idea how to design user interfaces...
Now that I'm all in the groove of CellML, I guess I should look into that API and things. I couldn't persuade it to run on my computer though, so we'll see how that works...
Oh yeah, I remember why I couldn't persuade it to work. It only compiles on Linux. Sigh. In my opinion, a program that can only compile in Linux is just as useless as one that can only compile in Windows. But that's probably my Java background talking.
Alright, back to SBW then. Hehe, the SBML support module is called NOM. NOM NOM NOM.
I asked Neil for the documents defining promoters and differeent parts, and he gave me several documents to read.
AGH.
[Error] ..\..\Users\Morgan\regulatory.cellml: the free variable '' is not defined anywhere
Why!
 "
"