|
Home
The Team
Project Report
Parts
Modeling
Notebook
Safety
CoLABoration
|
__october
Oct. 1st 2008
Digestion transfectionvector and CMV PCR product(3rd try)(Kathrin)
Digestion with EcoRI and PstI
Ligation transfectionvector and CMV PCR product(3rd try) (Kathrin)
Using new T4 DNA ligase and new ligase buffer
Miniprep of pBSK-signalpeptide (Sabine)
Transformation of pMA-transmembrane, pMA-Histag, pMA-signalpeptide, pMA-splitlinker, pMA-StreptagII (Sabine)
Oct. 2nd 2008
Picking clones (Kathrin)
pMA-transmembrane, pMA-Histag, pMA-signalpeptide, pMA-splitlinker, pMA-StreptagII
Digestion of: (Kathrin)
pBSK-signalpeptide with AgeI and PstI
pMA-Lipocalin and pMA-Anti-Nip-scFv with NgoMIV and PstI
Ligation (Kathrin)
Using Quick DNA ligase
Vector: pBSK-signalpeptide
Insert: Lipocalin, Anti-NIP-scFv
-Approaches: Insert/Vector -> a) 6/2, b) 4/4
Transformation (Kathrin)
of the ligation transfectionvector+CMV PCR product(3rd try)
Miniprep of: (Sabine)
pMA-transmembrane, pMA-Histag, pMA-signalpeptide, pMA-splitlinker, pMA-StreptagII
Preparative digestion of: (Sabine)
- pMA-splitlinker with AgeI and SpeI
- pMA-Cerulan split C-CFP and pMA-Venus split C-YFP with NgoMIV and SpeI
Ligation (Sabine)
Vector: pMA-splitlinker
Insert: Cerulan split C-CFP, Venus split C-YFP
Transformation (Sabine)
of the ligation pBSK-signalpeptide+Lipocalin and pBSK-signalpeptide+Anti-NIP-scFv
Oct. 3rd 2008
Transformation (Michael)
of the ligation pMA-splitlinker+Cerulan split C-CFP and pMA-splitlinker+Venus split C-YFP
Picking clones (Michael)
-transfectionvector-CMV PCR product
-pBSK-signalpeptide-Lipocalin
-pBSK-signalpeptide-Anti-NIP-scFv
Miniprep and analytic digestion of (Kathrin)
-transfectionvector-CMV PCR product
-pBSK-signalpeptide-Lipocalin
-pBSK-signalpeptide-Anti-NIP-scFv
Preparative digestion of (Kathrin)
-transfectionvector-CMV PCR product with SpeI and PstI
-pBSK-signalpeptide-Lipocalin and pBSK-signalpeptide-Anti-NIP-scFv with XbaI and PstI
Ligation (Sabine)
Vector: transfectionvector-CMV PCR product
Insert: signalpeptide-Lipocalin, signalpeptide-Anti-NIP-scFv
Oct. 4th 2008
Transformation (Normann)
-the ligation transfectionvector-CMV PCR product+signalpeptide-Lipocalin
-the ligation transfectionvector-CMV PCR product+signalpeptide-Anti-NIP-scFv
-pGA18-bla1(ß-Lactamase 1)
-pGA18-bla2(ß-Lactamase 2)
-pGA14-YFP
Picking clones (Sabine)
-pMA-splitlinker-Cerulan split C-CFP
-pMA-splitlinker-Venus split C-YFP
Oct. 5th 2008
Miniprep of: (Michael)
-pMA-splitlinker-Cerulan split C-CFP
-pMA-splitlinker-Venus split C-YFP
-pGA18-bla1
-pGA18-bla2
-pGA14-YFP
Picking clones (Sabine)
-transfectionvector-CMV PCR product-signalpeptide-Lipocalin
-transfectionvector-CMV PCR product-signalpeptide-Anti-NIP-scFv
Analytic digestion of: (Sabine)
-pMA-splitlinker-Cerulan split C-CFP -> 3 correct clones
-pMA-splitlinker-Venus split C-YFP -> 2 correct clones
Preparative digestion (Sabine)
-pGA14-YFP
Ligation (Sabine)
Using T4 DNA ligase
Vector: transfectionvector-CMV PCR product
Insert: YFP
Oct. 6th 2008
DNA-Origamis normann
DNA-Origamis with Alexa/noNIP, NIP/Alexa, NIP, noNIP were made.
The protocol of july 24th was used.
Preparation of 293T transfection normann
In order to transfect the 293T-cells with CMV+YFP tomorrow, the amount of cells per ml was determined with the "Neubauer cell chamber" --> 2,62*10E6 cells/ml
To transfect cells with 1µg DNA in a 6-well plate, you need ~6*10E4 cells per well, so 20µl of the suspension was given in each well and 2ml DMEM was added.
Transformation of the ligation (Sabine)
transfectionvector-CMV PCR product+YFP
Preparative digestion of (Sabine)
-pMA-transmembrane with AgeI and SpeI
-pMA-Cerulan N-CFP and pMA-Venus N-YFP and NgoMIV and SpeI
Preparative digestion of (Kathrin)
-pMA-splitlinker-Cerulan split C-CFP
-pMA-splitlinker-Venus split C-YFP
Analytic digestion of (Kathrin)
-transfectionvector-CMV PCR product-signalpeptide-Lipocalin -> no correct clones
-transfectionvector-CMV PCR product-signalpeptide-Anti-NIP-scFv -> no correct clones
Ligation (Michael)
Using Quick ligase
Vector: pMA-transmembrane
Insert: splitlinker-Cerulan split C-CFP, splitlinker-Venus split C-YFP, Cerulan N-CFP and Venus N-YFP
Transformation of the ligation (Michael)
-pMA-transmembrane+splitlinker-Cerulan split C-CFP
-pMA-transmembrane+splitlinker-Venus split C-YFP
-pMA-transmembrane+Cerulan N-CFP
-pMA-transmembrane+Venus N-YFP
Picking clones (Michael)
-transfectionvector-CMV PCR product-YFP
Oct. 7th 2008
Miniprep and analytic digestion (Kathrin)
- transfectionvector-CMV PCR product-YFP clones 1-4
- transfectionvector-CMV PCR product-Lipocalin clones 5-7
test digestion of the clones named above with NotI
analysis on the agarosegel shows not the expected bands
Oct. 8th 2008
Digestion of CFP normann
In order to put the gene for CFP behind the CMV-promotor on our transfectionvector to make a double trasnsfektion with YFP, the plasmit carrieng CFP was digested using pst1 and Xba1.
The digested gene was brought on an agarosegel and the expected band was cut out, after the followed the extraction out of the gel using the "QIAquick gel extraction kit"
preparation of ss Phage DNA normann
Because the ss PhageDNA ,which is needed for the origamis, is nearly empty phages were harvested out of a overnight culture.
Harvesting happened like it is descriped under Methods.
Preparation of the ssDNA was done following the QIAPrep Spin M13 Kits protocol.
Klenow-fill-in-reaction (Kathrin)
For producing a GGGS-Linker
Preparative digestion (Sabine)
-pMA-transmembrane with AgeI and SpeI
-pMA-BB058(Split luciferase), pGA18-bla1, pGA18-bla2 with NgoMIV and SpeI
Ligation (Sabine)
Using Quick ligase
Vector: pMA-transmembrane
Insert: BB058(Split luciferase), bla1 and bla2
Transformation (Sabine)
of the ligations
-pMA-transmembrane+BB058(Split luciferase)
-pMA-transmembrane+bla1
-pMA-transmembrane+bla2
Miniprep (Sabine)
-transfectionvector-CMV PCR product clones 5-9
Oct. 9th 2008
Analytic digestion (with NotI) (Kathrin)
-transfectionvector-CMV PCR product clones 5-9 -> correct clones
Preparative digestion (Kathrin)
-transfectionvector-CMV PCR product clone 5 with SpeI and PstI
-pBSK-signalpeptide-Lipocalin and pBSK-signalpeptide-Anti-Nip-sc-Fv with XbaI and PstI
Sequencing (Kathrin)
Correct:
-pMA-transmembrane-Cerulan-N-CFP clone 2
-pMA-transmembrane-Venus-N-YFP clone 3
-pMA-transmembrane-splitlinker-Cerulan-C-CFP clone 2
-pMA-transmembrane-splitlinker-Venus-C-YFP clone 3
Not correct:
-pBSK-signalpeptide-Lipocalin clone 1
-pBSK-signalpeptide-Anti-NIP-sc-Fv clone 4
Analytic Digestion (Sabine)
-pMA-signalpeptide-Anti-Nip-sc-Fv clones 1-3
-pMA-signalpeptide-Lipocalin clones 1-3
Preparative Digestion (Sabine)
-pMA-BB057(Splitluciferase) with NgoMIV and SpeI
-GGGS-Linker with NgoMIV and PstI
-pMA-signalpeptide-Anti-Nip-sc-Fv clone 2 with AgeI and PstI
-pMA-signalpeptide-Lipocalin clone3 with AgeI and PstI
Ligation (Sabine)
Using T4 DNA Ligase
Vector: pMA-signalpeptide-Anti-Nip-sc-Fv, pMA-signalpeptide-Lipocalin, pMA-transmembrane
Insert: GGGS-Linker, BB057
Oct. 10th 2008
Sequencing (Kathrin)
Correct:
-pMA-signalpeptide-Anti-Nip-sc-Fv clone 2
-pMA-signalpeptide-Lipocalin clone3
Transformation (Kathrin)
of the ligations
-pMA-signalpeptide-Anti-Nip-sc-Fv+GGGS-Linker
-pMA-signalpeptide-Lipocalin+GGGS-Linker
-pMA-transmembrane+BB057
Preparative Digestion (Sabine)
-pMA-transmembrane with AgeI and PstI
-BB057, BB058, bla1 and bla2 with NgoMIV and PstI
Picking clones (Sabine)
3 clones from pMA-signalpeptide-Lipocalin-GGGS-Linker
1 clone from pMA-signalpeptide-Anti-Nip-sc-Fv+GGGS-Linker
Oct. 11th 2008
Ligation of CFP/YFP and Transfectionvektor+CMV
Ligation was done using the quick ligase. Inkubation for 40min.
Transformation of TV-CMV-YFP and TV-CMV-CFP
Miniprep and analytic digestion(with NotI) (Sabine)
pMA-signalpeptide-Lipocalin-GGGS-Linker clones 1-3
Ligation (Sabine)
using the Quick ligation kit
Vector: pMA-transmembrane
Insert: BB057, BB058, bla1 and bla2
Preparative Digestion (Sabine)
-pMA-signalpeptide-Lipocalin-GGGS-Linker with AgeI and PstI
-pMA-transmembrane-Cerulan-N-CFP with NgoMIV and PstI
-pMA-transmembrane-Venus-N-YFP with NgoMIV and PstI
-pMA-transmembrane-splitlinker-Cerulan-C-CFP with NgoMIV and PstI
-pMA-transmembrane-splitlinker-Venus-C-YFP with NgoMIV and PstI
-> analysis on the agarosegel showed not the expected bands
Transformation (Michael)
of the ligations
pMA-transmembrane+BB057, pMA-transmembrane+BB058, pMA-transmembrane+bla1, pMA-transmembrane+bla2
Miniprep (Michael)
pMA-signalpeptide-Anti-Nip-sc-Fv+GGGS-Linker
Oct. 12th 2008
Miniprep (Michael)
pMA-transmembrane-Cerulan-N-CFP clone 2, pMA-transmembrane-Venus-N-YFP clone 3, pMA-transmembrane-splitlinker-Cerulan-C-CFP clone 2, pMA-transmembrane-splitlinker-Venus-C-YFP clone 3
Analytic Digestion (Kathrin)
With NotI
-pMA-signalpeptide-Lipocalin-GGGS-Linker clones 1-3(2nd try)
This time pMA-signalpeptide-Lipocalin was also digested to see the different sizes of the constructs with and without GGGS-Linker on the gel
-pMA-signalpeptide-Anti-Nip-sc-Fv+GGGS-Linker clone 1 and pMA-signalpeptide-Anti-Nip-sc-Fv
-> pMA-signalpeptide-Anti-Nip-sc-Fv+GGGS-Linker was not correct
Preparative Digestion (Kathrin)
-pMA-transmembrane-Cerulan-N-CFP clone 2, pMA-transmembrane-Venus-N-YFP clone 3, pMA-transmembrane-splitlinker-Cerulan-C-CFP clone 2 and pMA-transmembrane-splitlinker-Venus-C-YFP clone 3 with NgoMIV and PstI
-pMA-signalpeptide-Lipocalin-GGGS-Linker clone 3 with AgeI and PstI
-> not correct digested
-transfectionvector-CMV PCR product clone 5 with SpeI and PstI
- YFP clone 3, CFP clone 1 with XbaI and PstI
Ligation (Kathrin)
Using the Quick ligation kit
Vector: transfectionvector-CMV PCR product clone 5
Insert: YFP clone 3, CFP clone 1
Transformation (Sabine)
of the ligation transfectionvector-CMV PCR product clone 5+YFP clone 3 and transfectionvector-CMV PCR product clone 5+CFP clone 1
Oct. 13th 2008
Transformation (Kathrin)
repeat: transfectionvector-CMV PCR product clone 5+YFP clone 3 and +CFP clone 1
Miniprep (Kathrin)
- pMA-transmembraneregion-Splitlinker-C-Venus-YFP clone 3
- pMA-transmembraneregion-N-Venus-YFP clone 3
- pMA-transmembraneregion-Splitlinker-C-Cerulean-CFP clone 2
- pMA-transmembraneregion-N-Cerulean-CFP clone 2
Preparative Digestion (Kathrin)
for the final construct: pMA_Signalpeptide_scFv-anti-NIP_GGGS-Linker
digestion with AgeI and PstI: pMA_Signalpeptide_scFv-anti-NIP
digestion with NgoMIV and PstI: GGGS-linker (produced by fill in reaction)
for the final construct: pMA_Signalpeptide-Lipocalin_GGGS_N-CFP/ _N-YFP/ Splitlinker_C-CFP/ Splitlinker_C-YFP
digestion with AgeI and PstI: pMA_Signalpeptide-Lipocalin_GGGS-Linker clone 1
digestion with NgoMIV and PstI: pMA_transmembraneregion_N-CFP/ _N-YFP/ Splitlinker_C-CFP/ Splitlinker_C-YFP
Analytic Digestion (Kathrin)
the following constructs were tested: Transfecionvektor_CMV_clone5_YFP A, B, C, D
after gel-seperation the expected bands were detected (3400bp=vector + 1400bp=CMV+YFP)
clone D was chosen for a transfection of 293T-cells
20ml culture of Transfecionvektor_CMV_clone5_YFP D to get enough DNA for the transfection
Preparative Digestion (Michael)
for the final construct: pMA_transmembraneregion_Luciferase1/ _Luciferase2/ -ß-Lactamase1 /- ß-Lactamase2
vector-digestion with AgeI and PstI: pMA_transmembraneregion_clone3
insert-digestion with NgoMIV and PstI: pMA_BB058 (Luciferase1), pMA_BB057 (Luciferase2), pMA_bla1 (ß-Lactamase1), pMA_bla2 (ß-Lactamase2)
Ligation (Kathrin)
for the final constructs:
- pMA_Signalpeptide_scFv-anti-NIP_GGGS-Linker
- pMA_Signalpeptide-Lipocalin_GGGS_N-CFP
- pMA_Signalpeptide-Lipocalin_GGGS_N-YFP
- pMA_Signalpeptide-Lipocalin_GGGS_Splitlinker_C-CFP
- pMA_Signalpeptide-Lipocalin_GGGS_Splitlinker_C-YFP
Picking clones (Kathrin)
transfectionvector_CMV_PCR-product_clone5_YFP E, F, G, H
transfectionvector_CMV_PCR-product_clone5_CFP 1, 2, 3, 4
Ligation (Michael)
for the final constructs:
- pMA_transmembraneregion_Luciferase 1
- pMA_transmembraneregion_Luciferase 2
- pMA_transmembraneregion_ß-Lactamase1
- pMA_transmembraneregion_ß-Lactamase2
Transformation (Michael)
of all ligations mentioned above
Miniprep (Sabine)
pMA_BB057, _BB058, _transmembraneregion_clone3, _bla1, _bla2
Oct. 14th 2008
Miniprep (Kathrin)
- 16ml TV_CMV_clone5_YFP clone D for transfection of 293T cells
- 4ml TV_CMV_clone5_CFP clone 1, 2, 3, 4
- 4ml TV_CMV_clone5_YFP clone E, F, G, H
of the 4ml cultures also glycerinstockes were taken
Analytic Digestion (Kathrin)
- TV_CMV_clone5_CFP clone 1, 2, 3, 4
- TV_CMV_clone5_YFP clone E, F, G, H
expected bands were detected (3400bp=vector + 1400bp=CMV+YFP/ CMV+CFP)
Sequencing (Kathrin)
Correct:
- pMA_Signalpeptide_Lipocalin_GGGS-linker_clone 1
- pMA_Signalpeptide_Lipocalin_GGGS-linker_clone 2
Not correct
- TV_CMV_clone5_YFP clone A (base exchange in the CMV-promotor)
Picking clones (Michael)
- pMA_transmembraneregion_Luciferase 1
- pMA_transmembraneregion_Luciferase 2
- pMA_transmembraneregion_ß-Lactamase1
- pMA_transmembraneregion_ß-Lactamase2
- pMA_Signalpeptide_scFv-anti-NIP_GGGS-Linker
- pMA_Signalpeptide-Lipocalin_GGGS_N-CFP
- pMA_Signalpeptide-Lipocalin_GGGS_N-YFP
- pMA_Signalpeptide-Lipocalin_GGGS_Splitlinker_C-CFP
- pMA_Signalpeptide-Lipocalin_GGGS_Splitlinker_C-YFP
Oct. 15th 2008
Miniprep (Kathrin)
of the 18 clones picked the day before (mentioned above)
Analytic Digestion (Kathrin)
of the 18 clones picked the day before (mentioned above)
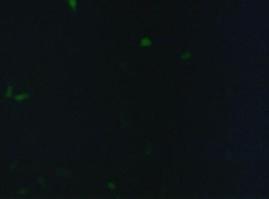
fluorescence microscopy (Normann and Kathrin)
to test if the CMV-promotor in the transfectionvector allows a succesful transfection of a protein TV_CMV_5_YFP clone D was transfected to 293T cells and analysed by fluorescence microscopy.
Preparative Digestion (Sabine)
for the final constructs: pMA_signalpeptide_scFv-antiNIP_GGGS-linker_transmembraneregion_bla1/ _bla2/ _Luciferase-BB058/ _Luciferase-BB057
digestion with AgeI and PstI: pMA_signalpeptide_scFv-antiNIP_GGGS-linker_
digestion with NgoMIV and PstI:pMA_transmembraneregion_bla1/ _bla2/ _Luciferase-BB058/ _Luciferase-BB057
Sequencing (Kathrin)
Correct:
- TV_CMV_clone 7
- TV_CMV_clone 8
Not correct
- TV_CMV_clone 5 (base exchange in the CMV promotor)
- TV_CMV_clone 9 (base exchange in the CMV promotor)
- TV_CMV_clone5_YFP A (base exchange in the CMV promotor)
- TV_CMV_clone5_CFP 1 (base exchange in the CMV promotor)
Ligation (Michael)
Using Quick ligase
Vector: pMA-signalpeptide-scFV-antiNIP-GGGSlinker
Insert: transmembrane-bla1, transmembrane-bla2, transmembrane-splitlinker-C-CFP, transmembrane-splitlinker-C-YFP, transmemrbane-N-CFP, transmembrane-N-YFP, transmembrane-Splitluciferase(BB058), transmembrane-Splitluciferase(BB057)
Vector: pMA-signalpeptide-Lipocalin-GGGSlinker
Insert: transmembrane-bla1, transmembrane-bla2, transmembrane-Splitluciferase(BB058), transmembrane-Splitluciferase(BB057)
Transformation (Michael)
of the ligations mentioned above
Oct. 16th 2008
Preparative Digestion (Sabine)
of Lipo C,D,E and F (always clone 1) with XbaI and PstI
to bring into the transfectionvector TV/CMV clone 7, digested with SpeI and PstI
Ligation (Michael)
of the digested vector and inserts (see above) with Quick ligase
Transformation (Michael)
of Lipo III, IV, V and VI in XL-1 cells
|