Origami-DNA
From 2008.igem.org
(Difference between revisions)
m |
|||
| Line 6: | Line 6: | ||
==Introduction== | ==Introduction== | ||
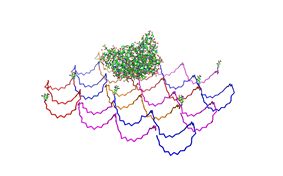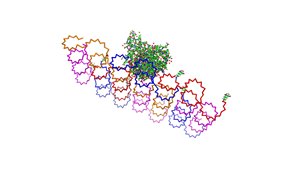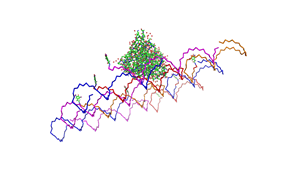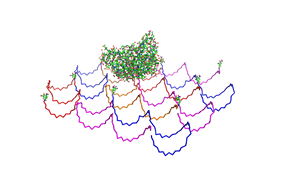
Paul Rothemund has discovered that it is possible to shape M13-Phage single-strand-DNA simply adding oligonucleotides that will work as „brackets“ when complementing the long single-strand.<br> In this way, one can generate for example DNA-squares of a certain size with „nods“ at certain distances. | Paul Rothemund has discovered that it is possible to shape M13-Phage single-strand-DNA simply adding oligonucleotides that will work as „brackets“ when complementing the long single-strand.<br> In this way, one can generate for example DNA-squares of a certain size with „nods“ at certain distances. | ||
| - | One member of our team, Daniel Hautzinger, has recently finished his diploma-thesis on Origami-DNA and the possibilities of generating patterns on these square surfaces by modifying the | + | One member of our team, Daniel Hautzinger, has recently finished his diploma-thesis on Origami-DNA and the possibilities of generating patterns on these square surfaces by modifying the oligo-nucleotides that build up the nod-points.<br> |
| + | As the antigens NIP and fluoresceine can as well be fused to these oligos, we had found the seemingly perfect tool to present strictly defined two-dimensional antigen-patterns to cells carrying our synthetic receptor system. | ||
[[Image:Freiburg2008_Fab_on_Origami_animated.gif|800 px]] | [[Image:Freiburg2008_Fab_on_Origami_animated.gif|800 px]] | ||
| + | |||
| + | ==Literature== | ||
| + | *Paul W. K. Rothemund: Nature 440, 297-302 (16 March 2006) | ||
}} | }} | ||
Revision as of 19:50, 23 October 2008
 "
"