Team:BrownTwo/Implementation/construction
From 2008.igem.org
| Line 1: | Line 1: | ||
{{limitertemp}} | {{limitertemp}} | ||
| - | == | + | ==A Modular Approach to Constructing the Circuit== |
| - | + | [[image:Limiter_diagram.PNG|center|600px]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | </ | + | |
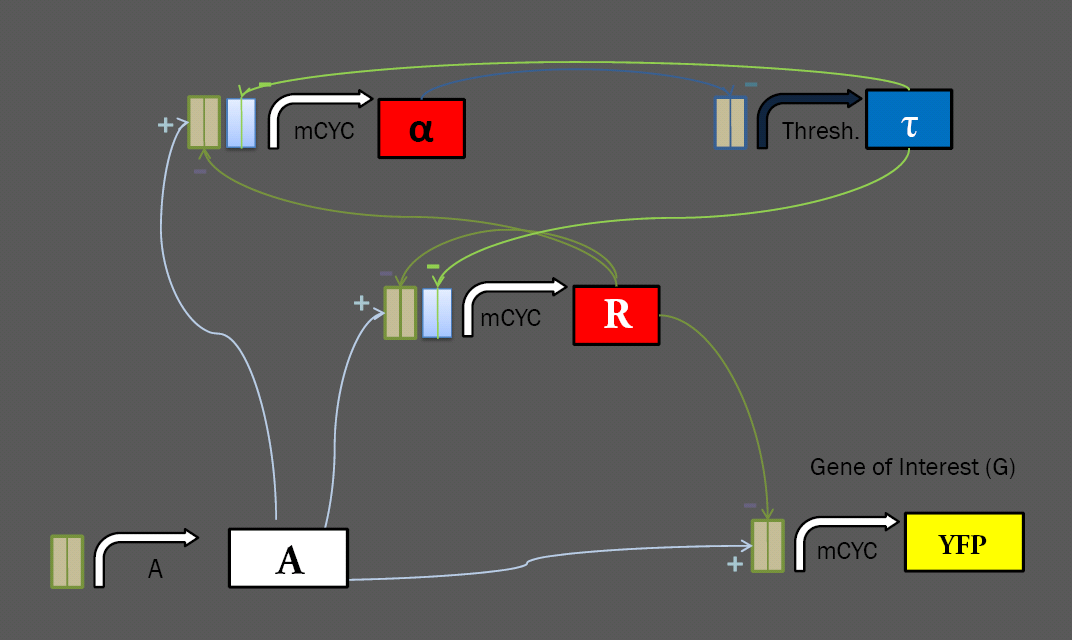
| + | <p>The general layout of our limiter circuit is very amenable to variation. Each of the logical roles can be filled by a number of different parts, possibly resulting in different behavior. This process was made especially straightforward by the modular transcriptional control elements at our disposal. Work with these transcription factors in the Silver Lab at Harvard showed that they show efficacy in regulating a variety of promoters, including the minimal mCYC1 and the inducible pGAL1. To extend this work further, we have constructed a library of regulable promoters for use in our network, ligating binding sites for the synthetic transcription factors onto a variety of yeast promoters.</p> | ||
| + | |||
| + | <p>mCYC was used as a minimal promoter, with all transcriptional induction and repression coming from transcription factors binding to sites ligated upstream. We generated a variety of minimal promoters for use in the limiter system, with mCYC ligated to each of the four sets of binding sites, and three variations on mCYC ligated to two different sets of binding sites. These will allow for construction of multiple variations on the limiter circuit. | ||
| + | </p> | ||
| + | |||
| + | <p>In a limiter circuit with a static threshold, the Thresh. promoter above is constitutive, but also requires regulation from alpha. To investigate the effect of changing the threshold, we ligated binding sites onto two constitutive promoters: the high-level TEF2, and the low-level ADH1.</p> | ||
| + | |||
| + | <p>Work in the Silver Lab has shown success in using binding sites to regulate inducible promoters. We have constructed two versions of the MET25 promoter with binding sites, hoping to construct a limiter circuit in which we can set the threshold by varying the input concentration of methionine. | ||
Revision as of 03:41, 30 October 2008
A Modular Approach to Constructing the Circuit
The general layout of our limiter circuit is very amenable to variation. Each of the logical roles can be filled by a number of different parts, possibly resulting in different behavior. This process was made especially straightforward by the modular transcriptional control elements at our disposal. Work with these transcription factors in the Silver Lab at Harvard showed that they show efficacy in regulating a variety of promoters, including the minimal mCYC1 and the inducible pGAL1. To extend this work further, we have constructed a library of regulable promoters for use in our network, ligating binding sites for the synthetic transcription factors onto a variety of yeast promoters. mCYC was used as a minimal promoter, with all transcriptional induction and repression coming from transcription factors binding to sites ligated upstream. We generated a variety of minimal promoters for use in the limiter system, with mCYC ligated to each of the four sets of binding sites, and three variations on mCYC ligated to two different sets of binding sites. These will allow for construction of multiple variations on the limiter circuit. In a limiter circuit with a static threshold, the Thresh. promoter above is constitutive, but also requires regulation from alpha. To investigate the effect of changing the threshold, we ligated binding sites onto two constitutive promoters: the high-level TEF2, and the low-level ADH1. Work in the Silver Lab has shown success in using binding sites to regulate inducible promoters. We have constructed two versions of the MET25 promoter with binding sites, hoping to construct a limiter circuit in which we can set the threshold by varying the input concentration of methionine. |
 "
"