Team:Chiba/Project
From 2008.igem.org
(→How Our System Works) |
(→What our system requires) |
||
| Line 81: | Line 81: | ||
| - | + | ||
| - | + | ||
| - | + | ||
===How Our System Works=== | ===How Our System Works=== | ||
Revision as of 05:44, 25 October 2008
| Home | The Team | The Project | Parts Submitted to the Registry | Reference | Notebook | Acknowledgements |
|---|
|
Project Design ( Sender experiments | Receiver experiments ) | Our Goal |
Introduction
- "Team : Chiba - E.coli time manager"
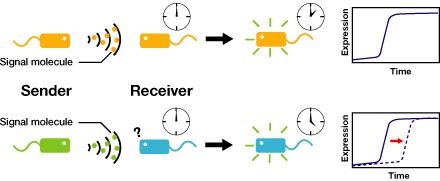
We control the timing of gene expression by using multiple signaling devices.To this end,we utilize molecules associated with Quorum sensing, a phenomenon that allows bacteria to communicate with each other.Our project uses two classes of bacteria: senders and receivers. Senders produce signaling molecules, and Receivers are activated only after a particular concentration of this molecule is reached.Although different quorum sensing species have slightly different signaling molecules, these molecules are not completely specific to their hosts and cross-species reactivity is observed [http://www3.interscience.wiley.com/journal/119124142/abstract (M.K Winson et al.FEMS Microbiology Letters,1998)]. Communication using non-endogenous molecules is less sensitive, and requires a higher signal concentration to take effect.This results in slower activation of receivers.
Motivation
Project Design
Our project uses two classes of bacteria: senders and receivers.Senders produce signaling molecules, and receivers are activated only after a particular concentration of this molecule is reached.The communication using non-endogenous molecules is less sensitive,and it requires higher signal concentration to take effect.This results in slower activation of receivers.
About Quorum Sensing
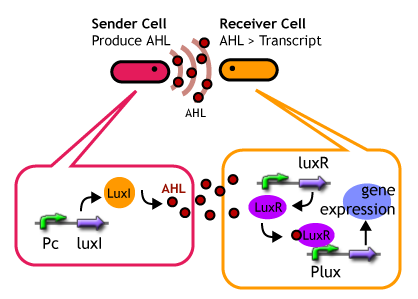
Quorum sensing is a cell-to-cell signaling action of bacteria. They detect the cell density of the same species and coordinate the expression behavior of their cells. Species of Gram-Negative signaling transfer molecules (so-called autoinducer) is a series of acyl homoserine lactone (AHL). The signals are synthesized from S-adenosylmethionine(SAM) by a synthase protein and once they have reached a threshold concentration,they bound to a transcriptional regulatory protein to induce expression of target genes.
More about Quorum Sensing
- [http://parts.mit.edu/registry/index.php/Featured_Parts:Cell-Cell-Signaling Cell-Cell-Signaling]
- [http://www.che.caltech.edu/groups/fha/quorum.html About Quorum sensing]
Controlling the time of a cell-to-cell signaling action
Communication using non-endogenous molecules is less sensitive, and requires a higher signal concentration to take effect.This results in slower activation of receivers.
- Sender Phase
- Quorum-Sensing Cross-talk
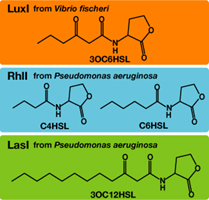
AHLs produced by different bacteria differ only in the length of the acyl-chain moiety and substitution at position C-3.Vibrio fischeri由来のLuxIは3OC6HSLを合成し、Pseudomonas aeruginosa由来のRhlIはC4HSLとC6HSLを合成し、LasIは3OC12HSLを合成する。
AHLを受け取り、遺伝子発現を活性化するLuxRはVibrio fischeri由来であり、3OC6HSLに対して低濃度でも、遺伝子発現が起こる。しかし、そのほかのAHLに対してはより高い濃度でなくては遺伝子発現が起こりにくいことが分かっている。 ([http://partsregistry.org/Part:BBa_F2620:Specificity BBa_F2620:Specificity])
AHLの濃度が上昇する速さが十分に遅いならば、遺伝子発現に時間差をつけることができるのではないかと私たちは考えた。
- Receiver Phase
AHLを合成するSenderだけではなく、AHLを受け取る側のReceiverにもさまざまな種類があれば、時間調節により幅を持たせることができる。そこで私たちは、いくつかの方法を考えた。
- 由来生物の違うレシーバータンパク質を発現させ、Receiver側でクロストークさせること
- レシーバータンパク質であるLuxRに変異を入れることで、AHLに対する応答感度を上下させること
- AHLを分解し、より多くのAHLが合成されなければ遺伝子発現が起こらないようにすること
- LuxR/Plux mutants show
- a greater response to 3OC6HSL ([http://authors.library.caltech.edu/5553/ C. H. Collins.et al.Mol.Microbiol.2005.])
- a increase in sensitivity to 3OC12HSL ([http://mic.sgmjournals.org/cgi/content/abstract/151/11/3589 B. Koch.et al.Microbiology (2005)]).
- AHL reporter with aiiA
- Express LuxR and aiiA constantly. AiiA degrades AHL as signaling molecule. Express GFP when the AHL concentration exceed the capacity of aiiA.
- This enables the delay of the activation time of receiver.
How Our System Works
| Home | The Team | The Project | Parts Submitted to the Registry | Reference | Notebook | Acknowledgements |
|---|
 "
"