Team:Davidson-Missouri Western
From 2008.igem.org
Macampbell (Talk | contribs) |
(→Highlights For Judges) |
||
| (51 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{| style="color:#1b2c8a;background-color:#0c6;" cellpadding="3" cellspacing="1" border="1" bordercolor="#fff" width="62%" align="center" | {| style="color:#1b2c8a;background-color:#0c6;" cellpadding="3" cellspacing="1" border="1" bordercolor="#fff" width="62%" align="center" | ||
| + | ![[Image:Spy1.jpg|30px]] <br> | ||
!align="center"|[[Team:Davidson-Missouri_Western|Home]] | !align="center"|[[Team:Davidson-Missouri_Western|Home]] | ||
!align="center"|[[Team:Davidson-Missouri_Western/Team|The Team]] | !align="center"|[[Team:Davidson-Missouri_Western/Team|The Team]] | ||
!align="center"|[[Team:Davidson-Missouri_Western/Project|''E. nigma'' Project]] | !align="center"|[[Team:Davidson-Missouri_Western/Project|''E. nigma'' Project]] | ||
!align="center"|[[Team:Davidson-Missouri_Western/New_Parts_Contributed_to_the_Registry|Parts Submitted to the Registry]] | !align="center"|[[Team:Davidson-Missouri_Western/New_Parts_Contributed_to_the_Registry|Parts Submitted to the Registry]] | ||
| - | |||
!align="center"|[[Team:Davidson-Missouri_Western/Notebook|Notebook]] | !align="center"|[[Team:Davidson-Missouri_Western/Notebook|Notebook]] | ||
|} | |} | ||
| Line 14: | Line 14: | ||
iGEM 2008 | iGEM 2008 | ||
</center></font> | </center></font> | ||
| - | [[Image: | + | <center> |
| + | [[Image:E_nigma_logo.jpg|400 px]] [[Image:vizabrick_logo.png|259 px]] | ||
| + | <br> | ||
| + | </center> | ||
| + | ''' ''E. nigma'': XOR Gates, a Bacterial Hash Function, and Viz-A-Brick''' | ||
| - | + | Our team designed, modeled, and constructed a bacterial computer that uses XOR logic to compute a cryptographic hash function. Hash functions are used to authenticate the integrity of a document by computing its digital fingerprint. Our bacterial computers are designed to recognize the presence or absence of two chemical signals via intercellular communication. Mathematical modeling of these computers has shown that our hash functions are difficult to corrupt. We designed and built a number of new parts, and improved and gained experience on existing parts. We also produced a graphical interface for exploring the Registry of Standard Biological Parts called Viz-A-Brick, and other web-based tools to improve the construction of new parts with BioBrick ends.<br><br> | |
| - | + | <center> | |
| - | Our team designed, modeled, and constructed a bacterial computer that uses XOR logic to compute a cryptographic hash function. Hash functions are used to authenticate the integrity of a document by computing its digital fingerprint. Our bacterial computers are designed to recognize the presence or absence of two chemical signals via intercellular communication. Mathematical modeling of these computers has shown that our hash functions are difficult to corrupt. We designed and built a number of new parts, and improved and gained experience on existing parts. We also produced a graphical interface for exploring the Registry of Standard Biological Parts called Viz-A-Brick, and other web-based tools to improve the construction of new parts with BioBrick ends. | + | <span style="color:red"> |
| - | + | ='''Highlights For Judges'''= | |
| - | + | </span> | |
| - | '''Highlights''' | + | </center> |
'''[[Team:Davidson-Missouri_Western/New Parts Contributed to the Registry|New Parts Contributed to the Registry]]''' - we designed, built, and contributed 105 new parts to the Registry | '''[[Team:Davidson-Missouri_Western/New Parts Contributed to the Registry|New Parts Contributed to the Registry]]''' - we designed, built, and contributed 105 new parts to the Registry | ||
| - | '''[[Team:Davidson-Missouri_Western/Experience Gained on New Registry Parts|Experience gained on new | + | '''[[Team:Davidson-Missouri_Western/Experience Gained on New Registry Parts|Experience gained on new Registry parts]]''' - we measured the function of several of the new parts we designed, and some expectations were met |
| - | '''[[Team:Davidson-Missouri_Western/Improvement of Pre-existing Registry Parts|Improvement of pre-existing | + | '''[[Team:Davidson-Missouri_Western/Improvement of Pre-existing Registry Parts|Improvement of pre-existing Registry parts]]''' - we redesigned the lac promoter and the lac repressor and measured an improvement in their functions |
| - | '''[[Team:Davidson-Missouri_Western/Online Tools that Support | + | '''[[Team:Davidson-Missouri_Western/Online Tools that Support Design of New Biobrick Parts|Online tools that support design of new BioBrick parts]]''' - we created online tools to aid in part design. |
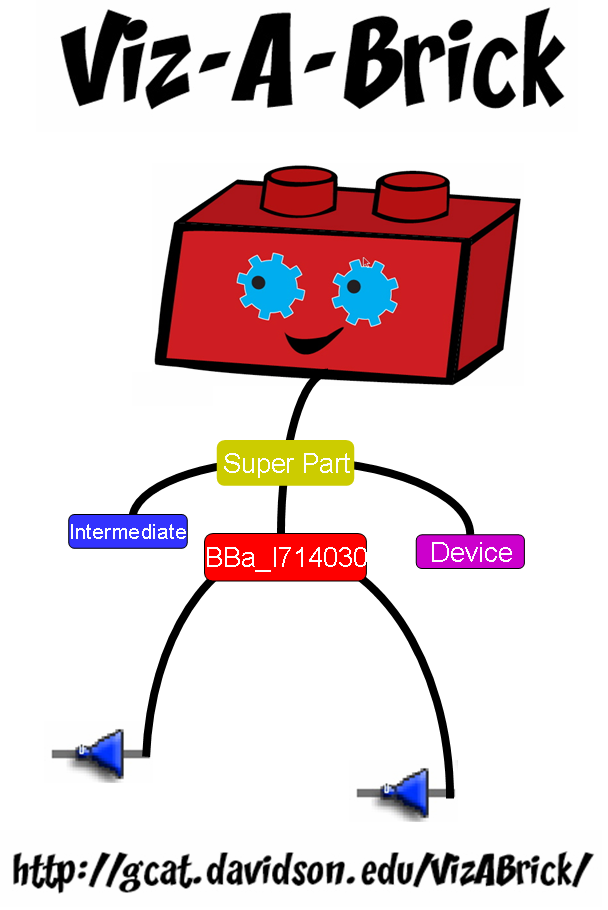
'''[[Team:Davidson-Missouri_Western/Viz-A-Brick: A New Way to Visualize the Registry|Viz-A-Brick: A new way to visualize the registry]]''' - we created a new interface to the Registry that addresses Human Practice issues associatated with navigating the expanding Registry | '''[[Team:Davidson-Missouri_Western/Viz-A-Brick: A New Way to Visualize the Registry|Viz-A-Brick: A new way to visualize the registry]]''' - we created a new interface to the Registry that addresses Human Practice issues associatated with navigating the expanding Registry | ||
| - | '''[[Team:Davidson-Missouri_Western/Project|E. nigma Project]]''' - we designed and constructed several systems for using logic gates to compute hash functions in bacteria | + | '''[[Team:Davidson-Missouri_Western/Project|''E. nigma'' Project]]''' - we designed and constructed several systems for using logic gates to compute hash functions in bacteria |
<br/> | <br/> | ||
| - | + | i'''GEM 2008 powerpoint presentation''' [[Media:MWSU_Davidson_iGEM08.PPT]] | |
| + | |||
| + | '''iGEM 2008 poster''' [[Media:MWSU_Davidson_iGEM08_poster.PPT]] | ||
| + | |||
| + | '''Funding''' - This collaborative project has received major support from National Science Foundation Grants #DMS-0733952 & 0733955, and was also supported by HHMI grant # 52006292, Davidson College DRI, the Davidson College Martin Genomics Program, the Missouri Western Foundation and the Missouri Western Office of Academic and Student Affairs. | ||
| - | + | <center> | |
| + | [[Image:Davidson_logo.JPG|140 px]] [[Image:Nsflogo.jpg|59 px]] [[Image:HHMI.gif]] [[Image:Western_logo.JPG|140 px]] | ||
| + | </center> | ||
| Line 49: | Line 59: | ||
!align="center"|[[Team:Davidson-Missouri_Western/Project|''E. nigma'' Project]] | !align="center"|[[Team:Davidson-Missouri_Western/Project|''E. nigma'' Project]] | ||
!align="center"|[[Team:Davidson-Missouri_Western/New_Parts_Contributed_to_the_Registry|Parts Submitted to the Registry]] | !align="center"|[[Team:Davidson-Missouri_Western/New_Parts_Contributed_to_the_Registry|Parts Submitted to the Registry]] | ||
| - | |||
!align="center"|[[Team:Davidson-Missouri_Western/Notebook|Notebook]] | !align="center"|[[Team:Davidson-Missouri_Western/Notebook|Notebook]] | ||
|} | |} | ||
Latest revision as of 03:55, 30 October 2008
| Home | The Team | E. nigma Project | Parts Submitted to the Registry | Notebook |
|---|
Davidson College - Missouri Western State University
iGEM 2008
E. nigma: XOR Gates, a Bacterial Hash Function, and Viz-A-Brick
Our team designed, modeled, and constructed a bacterial computer that uses XOR logic to compute a cryptographic hash function. Hash functions are used to authenticate the integrity of a document by computing its digital fingerprint. Our bacterial computers are designed to recognize the presence or absence of two chemical signals via intercellular communication. Mathematical modeling of these computers has shown that our hash functions are difficult to corrupt. We designed and built a number of new parts, and improved and gained experience on existing parts. We also produced a graphical interface for exploring the Registry of Standard Biological Parts called Viz-A-Brick, and other web-based tools to improve the construction of new parts with BioBrick ends.
Highlights For Judges
New Parts Contributed to the Registry - we designed, built, and contributed 105 new parts to the Registry
Experience gained on new Registry parts - we measured the function of several of the new parts we designed, and some expectations were met
Improvement of pre-existing Registry parts - we redesigned the lac promoter and the lac repressor and measured an improvement in their functions
Online tools that support design of new BioBrick parts - we created online tools to aid in part design.
Viz-A-Brick: A new way to visualize the registry - we created a new interface to the Registry that addresses Human Practice issues associatated with navigating the expanding Registry
E. nigma Project - we designed and constructed several systems for using logic gates to compute hash functions in bacteria
iGEM 2008 powerpoint presentation Media:MWSU_Davidson_iGEM08.PPT
iGEM 2008 poster Media:MWSU_Davidson_iGEM08_poster.PPT
Funding - This collaborative project has received major support from National Science Foundation Grants #DMS-0733952 & 0733955, and was also supported by HHMI grant # 52006292, Davidson College DRI, the Davidson College Martin Genomics Program, the Missouri Western Foundation and the Missouri Western Office of Academic and Student Affairs.
| Home | The Team | E. nigma Project | Parts Submitted to the Registry | Notebook |
|---|
 "
"