Team:Davidson-Missouri Western/Online Tools that Support Design of New Biobrick Parts
From 2008.igem.org
Macampbell (Talk | contribs) |
Macampbell (Talk | contribs) |
||
| (5 intermediate revisions not shown) | |||
| Line 7: | Line 7: | ||
|} | |} | ||
| - | == | + | == We recognized needs of the iGEM community and produced online tools to address some of these.== |
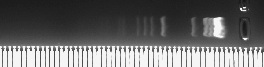
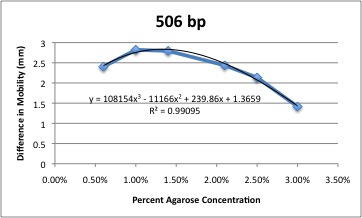
| - | 1. [http://gcat.davidson.edu/iGEM08/gelwebsite/gelwebsite.html '''Optimal Percent Agarose Gel''']<br> | + | 1. [http://gcat.davidson.edu/iGEM08/gelwebsite/gelwebsite.html '''Optimal Percent Agarose Gel''' (iGEM 2008)]<br> |
| - | How do you choose the right percent gel for separating any two DNA bands? Rather than guessing, there | + | How do you choose the right percent gel for separating any two DNA bands? Rather than guessing, there needs to be a more reliable method, especially since so many iGEM students are new to the techniques of the lab. Therefore, we produced a Gel Optimizer to help choose the right percent agarose gel. When you submit the MW of your band, you will get the optimal percent agarose (between 2% and 0.4%) as well as a photo showing molecular weight markers at this percent gel and the equation used to determine the migration of the nearest MW marker fragment at the different gel concentrations. <br> |
<center> | <center> | ||
[[Image:point8_gel.jpg]]<br> | [[Image:point8_gel.jpg]]<br> | ||
| Line 18: | Line 18: | ||
| - | 2. [http://gcat.davidson.edu/iGEM08/bbprimer.html '''PCR Primers with Digestable BioBrick Ends''']<br> | + | 2. [http://gcat.davidson.edu/iGEM08/bbprimer.html '''PCR Primers with Digestable BioBrick Ends''' (iGEM 2008)]<br> |
| - | Can we design PCR primers that include BioBrick ends automatically? We | + | Can we design PCR primers that include BioBrick ends automatically? We generated a webpage that allows users to submit the fragment of DNA they want amplified and then produce synthesis-ready PCR primers that not only amplify the fragment, but append the BioBrick prefix and suffix nucleotides as well as 4 additional bases (GCAT) so you can digest the PCR product with EcoRI and PstI and generate sticky ends. The primers appear in blue with the four BioBrick restriction sites underlined. The additional GCAT bases appear red. <br> |
<center> | <center> | ||
[[Image:Primer_ends.jpg]] | [[Image:Primer_ends.jpg]] | ||
</center> | </center> | ||
| - | |||
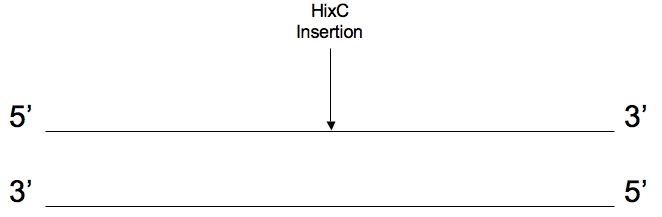
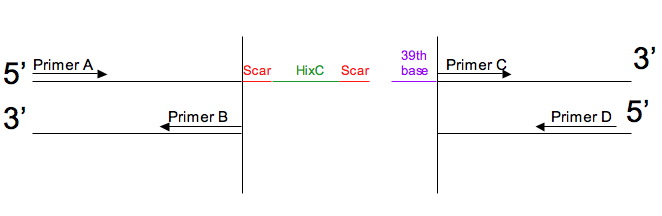
| - | + | 3. [http://gcat.davidson.edu/iGEM07/genesplitter.html '''Splitting genes for Hin/Hix flipping''' (iGEM 2007)]<br> | |
| - | + | This page is designed to tell you what 4 PCR primers can be used to split a gene so that hixC can be inserted into the gene's coding sequence and not alter the reading frame. <br> | |
| - | + | ||
| - | + | ||
| - | 3. [http://gcat.davidson.edu/iGEM07/genesplitter.html '''Splitting genes for Hin/Hix flipping''' (iGEM 2007)] | + | |
| - | This page is designed to tell you what 4 PCR primers can be used to split a gene | + | |
<center> | <center> | ||
'''Target Site'''<br> | '''Target Site'''<br> | ||
| Line 39: | Line 34: | ||
</center> | </center> | ||
| - | 4. [http://gcat.davidson.edu/IGEM06/oligo.html '''Do-It-Yourself Gene Assembly: Gene Synthesis Optimization Program''' | + | 4. [http://gcat.davidson.edu/IGEM06/oligo.html '''Do-It-Yourself Gene Assembly: Gene Synthesis Optimization Program''' (iGEM 2006)] <br> |
| - | This web tool allows you to submit any sequence under 300 bp and have it produce for you the oligos you need synthesized to self-assemble the entire dsDNA piece. It includes the BioBrick ends, but you will need to manually trim off 4 bases on each end to produce EcoRI and PstI sticky ends once the DNA is assembled. The program automatically optimizes oligos of similar melting temperature so all you have to do is mix, boil, cool, ligate. <br> | + | This web tool allows you to submit any sequence under 300 bp and have it produce for you the oligos you need synthesized to self-assemble the entire dsDNA piece. It includes the BioBrick ends, but you will need to manually trim off 4 bases on each end to produce EcoRI and PstI sticky ends once the DNA is assembled. The program automatically optimizes oligos of similar melting temperature so all you have to do is mix, boil, cool, and ligate. <br> |
<center> | <center> | ||
[[Image:Assembly_site.jpg]] | [[Image:Assembly_site.jpg]] | ||
Latest revision as of 20:28, 29 October 2008
| Home | The Team | E. nigma Project | Parts Submitted to the Registry | Notebook |
|---|
We recognized needs of the iGEM community and produced online tools to address some of these.
1. Optimal Percent Agarose Gel (iGEM 2008)
How do you choose the right percent gel for separating any two DNA bands? Rather than guessing, there needs to be a more reliable method, especially since so many iGEM students are new to the techniques of the lab. Therefore, we produced a Gel Optimizer to help choose the right percent agarose gel. When you submit the MW of your band, you will get the optimal percent agarose (between 2% and 0.4%) as well as a photo showing molecular weight markers at this percent gel and the equation used to determine the migration of the nearest MW marker fragment at the different gel concentrations.
2. PCR Primers with Digestable BioBrick Ends (iGEM 2008)
Can we design PCR primers that include BioBrick ends automatically? We generated a webpage that allows users to submit the fragment of DNA they want amplified and then produce synthesis-ready PCR primers that not only amplify the fragment, but append the BioBrick prefix and suffix nucleotides as well as 4 additional bases (GCAT) so you can digest the PCR product with EcoRI and PstI and generate sticky ends. The primers appear in blue with the four BioBrick restriction sites underlined. The additional GCAT bases appear red.
3. Splitting genes for Hin/Hix flipping (iGEM 2007)
This page is designed to tell you what 4 PCR primers can be used to split a gene so that hixC can be inserted into the gene's coding sequence and not alter the reading frame.
4. Do-It-Yourself Gene Assembly: Gene Synthesis Optimization Program (iGEM 2006)
This web tool allows you to submit any sequence under 300 bp and have it produce for you the oligos you need synthesized to self-assemble the entire dsDNA piece. It includes the BioBrick ends, but you will need to manually trim off 4 bases on each end to produce EcoRI and PstI sticky ends once the DNA is assembled. The program automatically optimizes oligos of similar melting temperature so all you have to do is mix, boil, cool, and ligate.
| Home | The Team | E. nigma Project | Parts Submitted to the Registry | Notebook |
|---|
 "
"