Team:Davidson-Missouri Western/Project
From 2008.igem.org
(→Overall project) |
(→XOR Gate Blueprints) |
||
| (57 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | <!-- | + | {| style="color:#1b2c8a;background-color:#0c6;" cellpadding="3" cellspacing="1" border="1" bordercolor="#fff" width="62%" align="center" |
| + | !align="center"|[[Team:Davidson-Missouri_Western|Home]] | ||
| + | !align="center"|[[Team:Davidson-Missouri_Western/Team|The Team]] | ||
| + | ![[Image:Spy1.jpg|30px]] <br> | ||
| + | !align="center"|[[Team:Davidson-Missouri_Western/Project|''E. nigma'' Project]] | ||
| + | !align="center"|[[Team:Davidson-Missouri_Western/New_Parts_Contributed_to_the_Registry|Parts Submitted to the Registry]] | ||
| + | !align="center"|[[Team:Davidson-Missouri_Western/Notebook|Notebook]] | ||
| + | |} | ||
| - | + | ==''' ''E. nigma'' Project Overview: '''Using E. coli to compute values of a cryptographic hash function'''== | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | A recent article serves as a [http://gcat.davidson.edu/iGEM08/cryptography_graph.pdf International Call for a Better Hash Function], an algorithm that produces a digital fingerprint of a digitized message. We decided to work toward the design and construction of a bacterial hash function. To this end, we designed and constructed several novel dually-regulated hybrid promoters, crucial new elements in the genetic circuitry we designed to function as biological XOR gates. These gates produce a positive result in the presence of exactly one input and a negative result otherwise and can be put in sequence to create a bacterial hash function. The name of the project is a play on the name of the World War II coding machine used to encrypt military secrets. | |
| + | Our multidisciplinary team conducted a project that drew expertise from biology and mathematics to explore the possibility of designing, modeling, constructing, and testing logic gates that would enable bacteria to compute a hash function. The links below provide documentation of the diverse outcomes of our research, illustrating not only the feasibility of bacterial computation but the ability of undergraduates students to contribute to an important emerging field. | ||
| + | =='''Cryptographic Hash Functions'''== | ||
| - | + | A cryptographic hash function takes as input a message or document of any size, and returns a fixed length hexadecimal string as output, called the '''hash value'''. The current widely-held standard is called '''MD-5'''. The hash value is essentially the "digital signature" of the input document, and can be used in many cases to determine if a document has been tampered with. The hash function should be sensitive to small perturbations in the input message, producing very different hash values for highly similar, but not identical, documents. To learn more about the properties of hash functions, their applications, our hash function models, and our biological implementation of hash functions, follow the links below. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | '' | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | |||
{| style="color:#1b2c8a;background-color:#0c6;" cellpadding="3" cellspacing="1" border="1" bordercolor="#fff" width="62%" align="center" | {| style="color:#1b2c8a;background-color:#0c6;" cellpadding="3" cellspacing="1" border="1" bordercolor="#fff" width="62%" align="center" | ||
| - | !align="center"|[[Team:Davidson-Missouri_Western| | + | !align="center"|[[Team:Davidson-Missouri_Western/Ideal Hash Function Characteristics|Properties ]] |
| - | !align="center"|[[Team:Davidson-Missouri_Western/ | + | !align="center"|[[Team:Davidson-Missouri_Western/Applications of Hash Functions|Applications]] |
| - | !align="center"|[[Team:Davidson-Missouri_Western/ | + | !align="center"|[[Team:Davidson-Missouri_Western/XOR and Binary Messages|Binary Messages and XOR Logic]] |
| - | !align="center"|[[Team:Davidson-Missouri_Western/ | + | !align="center"|[[Team:Davidson-Missouri_Western/Our Models|Our Models]] |
| - | !align="center"|[[Team:Davidson-Missouri_Western/ | + | !align="center"|[[Team:Davidson-Missouri_Western/Analysis of Our Models|Analysis of our Models]] |
| - | !align="center"|[[Team:Davidson-Missouri_Western/ | + | !align="center"|[[Team:Davidson-Missouri_Western/Matlab Files|Matlab files]] |
| + | !align="center"|[[Team:Davidson-Missouri_Western/Future Work|Future work]] | ||
|} | |} | ||
| - | |||
| + | ==''' XOR Gate Blueprints '''== | ||
| + | Our team designed five distinct XOR gates based on three different, known, intercellular communication systems. An XOR gate receives some subset of two inputs and produces a strong signal precisely when exactly one of the inputs is present. While the design of logic gates has become prevalent among iGEM teams, to date, we are unaware of any successful XOR gate built from genetic circuitry. The link below leads to detailed information about the design and construction of our XOR gates. | ||
| - | + | <center>'''Two of the XOR gate designs are shown below'''</center> | |
| + | <br> | ||
| - | + | <center> [[Image:XOR_AMC1b.jpg|420 px]] [[Image:XOR DR AI2.PNG|420 px]] </center> | |
| - | + | <br> | |
| + | <center> '''Detailed descriptions of all five gates can be found here: [[Team:Davidson-Missouri_Western/DNA Encoded XOR Gates|DNA Encoded XOR Gates]]'''</center> | ||
| + | <br> | ||
| + | <br> | ||
| + | =='''Intercellular Communication Systems '''== | ||
| + | The ''E. nigma'' Project relies on the ability of one cell to communicate with another cell through distance in the form of a chemical signal. Several intercellular communication systems exist in nature. The cellular communications systems used in our project were quorum sensing systems found in the bacterial species ''Vibrio fischeri'', ''E. coli'', and ''Pseudomonas aeruginosa''. The links below provide information about the three communications systems, the signaling molecules used, the constructs we tested, and issues with delayed growth. | ||
| + | {| style="color:#1b2c8a;background-color:#0c6;" cellpadding="3" cellspacing="1" border="1" bordercolor="#fff" width="62%" align="center" | ||
| + | !align="center"|[[Cellular Communication Systems]] | ||
| + | !align="center"|[[Team:Davidson-Missouri_Western/XOR and Autoinducers|Signaling Molecules]] | ||
| + | !align="center"|[[Constructs tested]] | ||
| + | !align="center"|[[Team:Davidson-Missouri_Western/Time-Delayed Growth|Time-Delayed Growth]] ([http://www.bio.davidson.edu/courses/genomics/2008/DeLoache/TimeDelayedWithTimes.mov See the QT Movie]) | ||
| + | |} | ||
| - | |||
| + | =='''Parts Designed, Constructed, and Contributed'''== | ||
| + | Our team designed and synthesized several novel dually-regulated promoters for use in our XOR gate constructions. These promoters are among the 105 parts contributed to the Registry. | ||
| + | [[Team:Davidson-Missouri_Western/Hybrid Promoters|Hybrid Promoters]] | ||
| + | [http://partsregistry.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2008&group=Davidson-Missouri_Western parts contributed ] | ||
| + | [[Systems for sending and receiving]] | ||
| - | + | [[Team:Davidson-Missouri_Western/Experimental Data on XOR Gate|Experimental data on XOR gate]] | |
| + | <br> | ||
| + | =='''[[Team:Davidson-Missouri_Western/Viz-A-Brick: A New Way to Visualize the Registry|Viz-A-Brick: A new way to visualize the registry]]'''== | ||
| + | We created a new interface to the Registry that addresses Human Practice issues associatated with navigating the expanding Registry | ||
| - | |||
| - | == | + | {| style="color:#1b2c8a;background-color:#0c6;" cellpadding="3" cellspacing="1" border="1" bordercolor="#fff" width="62%" align="center" |
| + | !align="center"|[[Team:Davidson-Missouri_Western|Home]] | ||
| + | !align="center"|[[Team:Davidson-Missouri_Western/Team|The Team]] | ||
| + | !align="center"|[[Team:Davidson-Missouri_Western/Project|''E. nigma'' Project]] | ||
| + | !align="center"|[[Team:Davidson-Missouri_Western/New_Parts_Contributed_to_the_Registry|Parts Submitted to the Registry]] | ||
| + | !align="center"|[[Team:Davidson-Missouri_Western/Notebook|Notebook]] | ||
| + | |} | ||
Latest revision as of 03:49, 30 October 2008
| Home | The Team | E. nigma Project | Parts Submitted to the Registry | Notebook |
|---|
Contents |
E. nigma Project Overview: Using E. coli to compute values of a cryptographic hash function
A recent article serves as a [http://gcat.davidson.edu/iGEM08/cryptography_graph.pdf International Call for a Better Hash Function], an algorithm that produces a digital fingerprint of a digitized message. We decided to work toward the design and construction of a bacterial hash function. To this end, we designed and constructed several novel dually-regulated hybrid promoters, crucial new elements in the genetic circuitry we designed to function as biological XOR gates. These gates produce a positive result in the presence of exactly one input and a negative result otherwise and can be put in sequence to create a bacterial hash function. The name of the project is a play on the name of the World War II coding machine used to encrypt military secrets.
Our multidisciplinary team conducted a project that drew expertise from biology and mathematics to explore the possibility of designing, modeling, constructing, and testing logic gates that would enable bacteria to compute a hash function. The links below provide documentation of the diverse outcomes of our research, illustrating not only the feasibility of bacterial computation but the ability of undergraduates students to contribute to an important emerging field.
Cryptographic Hash Functions
A cryptographic hash function takes as input a message or document of any size, and returns a fixed length hexadecimal string as output, called the hash value. The current widely-held standard is called MD-5. The hash value is essentially the "digital signature" of the input document, and can be used in many cases to determine if a document has been tampered with. The hash function should be sensitive to small perturbations in the input message, producing very different hash values for highly similar, but not identical, documents. To learn more about the properties of hash functions, their applications, our hash function models, and our biological implementation of hash functions, follow the links below.
| Properties | Applications | Binary Messages and XOR Logic | Our Models | Analysis of our Models | Matlab files | Future work |
|---|
XOR Gate Blueprints
Our team designed five distinct XOR gates based on three different, known, intercellular communication systems. An XOR gate receives some subset of two inputs and produces a strong signal precisely when exactly one of the inputs is present. While the design of logic gates has become prevalent among iGEM teams, to date, we are unaware of any successful XOR gate built from genetic circuitry. The link below leads to detailed information about the design and construction of our XOR gates.
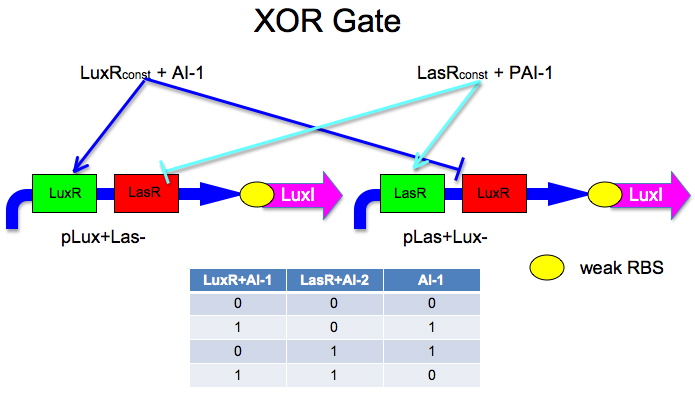
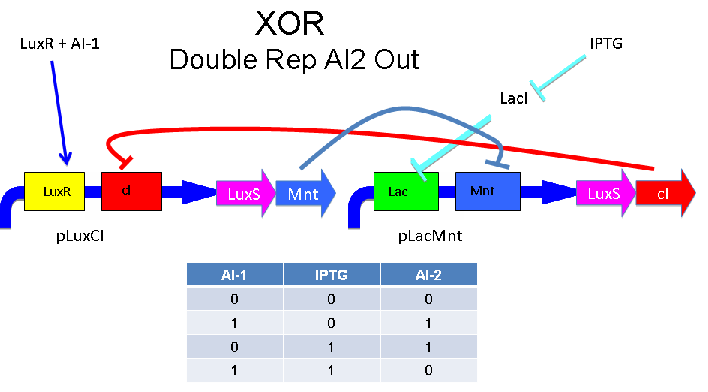
Intercellular Communication Systems
The E. nigma Project relies on the ability of one cell to communicate with another cell through distance in the form of a chemical signal. Several intercellular communication systems exist in nature. The cellular communications systems used in our project were quorum sensing systems found in the bacterial species Vibrio fischeri, E. coli, and Pseudomonas aeruginosa. The links below provide information about the three communications systems, the signaling molecules used, the constructs we tested, and issues with delayed growth.
| Cellular Communication Systems | Signaling Molecules | Constructs tested | Time-Delayed Growth ([http://www.bio.davidson.edu/courses/genomics/2008/DeLoache/TimeDelayedWithTimes.mov See the QT Movie]) |
|---|
Parts Designed, Constructed, and Contributed
Our team designed and synthesized several novel dually-regulated promoters for use in our XOR gate constructions. These promoters are among the 105 parts contributed to the Registry.
[http://partsregistry.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2008&group=Davidson-Missouri_Western parts contributed ]
Systems for sending and receiving
Viz-A-Brick: A new way to visualize the registry
We created a new interface to the Registry that addresses Human Practice issues associatated with navigating the expanding Registry
| Home | The Team | E. nigma Project | Parts Submitted to the Registry | Notebook |
|---|
 "
"