Team:EPF-Lausanne/2-step PCR
From 2008.igem.org
(→Time saved) |
|||
| (20 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{EPFL/Header}} | {{EPFL/Header}} | ||
| - | ==2- | + | ==2-step PCR== |
| - | The 2- | + | The 2-step PCR is a technique used to add small DNA sequences to a PCR product. In the context of iGEM, this can be used as an alternative to classical cloning: instead of digesting and ligating each biobrick, small parts can be directly added by PCR, saving a lot of time. |
| - | The 2-step PCR requires three pairs of | + | The 2-step PCR requires three pairs of primers and three PCR runs: |
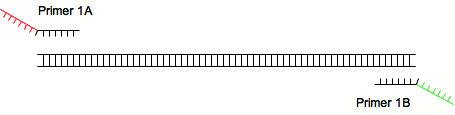
| - | A first pair is used to amplify a sequence of interest (for example an ORF) and provides annealing sites for the second pair of primers | + | A first pair is used to amplify a sequence of interest (for example an ORF) and provides annealing sites for the second pair of primers: |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
[[Image:2step1.png|center]] | [[Image:2step1.png|center]] | ||
| + | |||
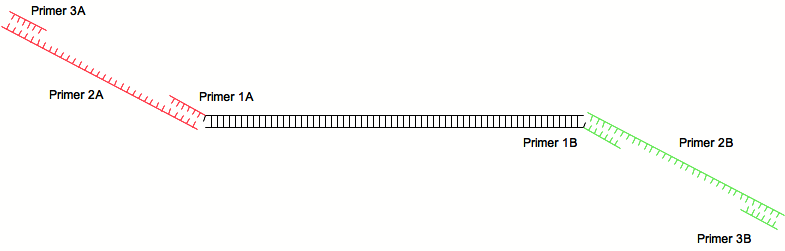
| + | The second pair anneals with the first and contains the sequence we want to add to the PCR product. The third pair of primers finally amplifies the initial PCR product along with the added sequence. | ||
[[Image:2step2.png|center]] | [[Image:2step2.png|center]] | ||
| + | |||
| + | Results: | ||
| + | |||
[[Image:2step3.png|center]] | [[Image:2step3.png|center]] | ||
| - | During our project, we used this technique to clone the ORF of the gene luxI | + | |
| + | During our project, we used this technique to clone the [https://2008.igem.org/wiki/index.php?title=EPF-Lausanne/29_September_2008 ORF of the gene ''luxI''] (the corresponding Biobrick, F1610, seemed to be inconsistent). The first PCR was performed on a plasmid to amplify the ORF coding for LuxI. During the second step, an RBS and the prefix were added upstream, whereas a terminator and the suffix were added downstream. The result of the this 2-step PCR was digested and inserted in pSB1A2, resulting in the Biobrick BBa_K092400. We are awaiting results on the generated sequence quality of this procedure. | ||
==Time saved== | ==Time saved== | ||
| - | The major advantage of this technique is time needed to create | + | The major advantage of this technique is that the time needed to create clones can be reduced significantly. A promoter, an RBS and a terminator can be added to an ORF in a few hours. The result of this 2-step PCR can be digested and ligated into a vector. In total, this operation takes approximately 30 hours. In comparison, generating the same clone using classical cloning methods would take six days. |
| - | + | ||
| - | + | ||
==A primer library?== | ==A primer library?== | ||
| + | |||
| + | Several cloning steps can be considered as being standard in an iGEM project. Adding RBS, promoter or terminator to an ORF is a common feature of almost all projects. For this reason, we suggest that a library containing primers designed for 2-step PCR should be created. This would give an easier and quicker way to assemble components. The new parts created with this technique keep all the Biobricks properties, meaning that classical cloning can also be used afterwards to assemble big parts together. | ||
| + | |||
| + | {{EPFL/Header}} | ||
Latest revision as of 17:14, 29 October 2008
| Home | The Team | The Project | Parts | 2-step PCR | Microfluidics | Modeling | Notebook |
|---|
2-step PCR
The 2-step PCR is a technique used to add small DNA sequences to a PCR product. In the context of iGEM, this can be used as an alternative to classical cloning: instead of digesting and ligating each biobrick, small parts can be directly added by PCR, saving a lot of time.
The 2-step PCR requires three pairs of primers and three PCR runs:
A first pair is used to amplify a sequence of interest (for example an ORF) and provides annealing sites for the second pair of primers:
The second pair anneals with the first and contains the sequence we want to add to the PCR product. The third pair of primers finally amplifies the initial PCR product along with the added sequence.
Results:
During our project, we used this technique to clone the ORF of the gene luxI (the corresponding Biobrick, F1610, seemed to be inconsistent). The first PCR was performed on a plasmid to amplify the ORF coding for LuxI. During the second step, an RBS and the prefix were added upstream, whereas a terminator and the suffix were added downstream. The result of the this 2-step PCR was digested and inserted in pSB1A2, resulting in the Biobrick BBa_K092400. We are awaiting results on the generated sequence quality of this procedure.
Time saved
The major advantage of this technique is that the time needed to create clones can be reduced significantly. A promoter, an RBS and a terminator can be added to an ORF in a few hours. The result of this 2-step PCR can be digested and ligated into a vector. In total, this operation takes approximately 30 hours. In comparison, generating the same clone using classical cloning methods would take six days.
A primer library?
Several cloning steps can be considered as being standard in an iGEM project. Adding RBS, promoter or terminator to an ORF is a common feature of almost all projects. For this reason, we suggest that a library containing primers designed for 2-step PCR should be created. This would give an easier and quicker way to assemble components. The new parts created with this technique keep all the Biobricks properties, meaning that classical cloning can also be used afterwards to assemble big parts together.
| Home | The Team | The Project | Parts | 2-step PCR | Microfluidics | Modeling | Notebook |
|---|
 "
"