Team:Hawaii/Notebook/2008-07-28
From 2008.igem.org
(Difference between revisions)
(→Transformation) |
(→Colony PCR) |
||
| Line 12: | Line 12: | ||
===Colony PCR=== | ===Colony PCR=== | ||
:<strong> Margaret</strong> | :<strong> Margaret</strong> | ||
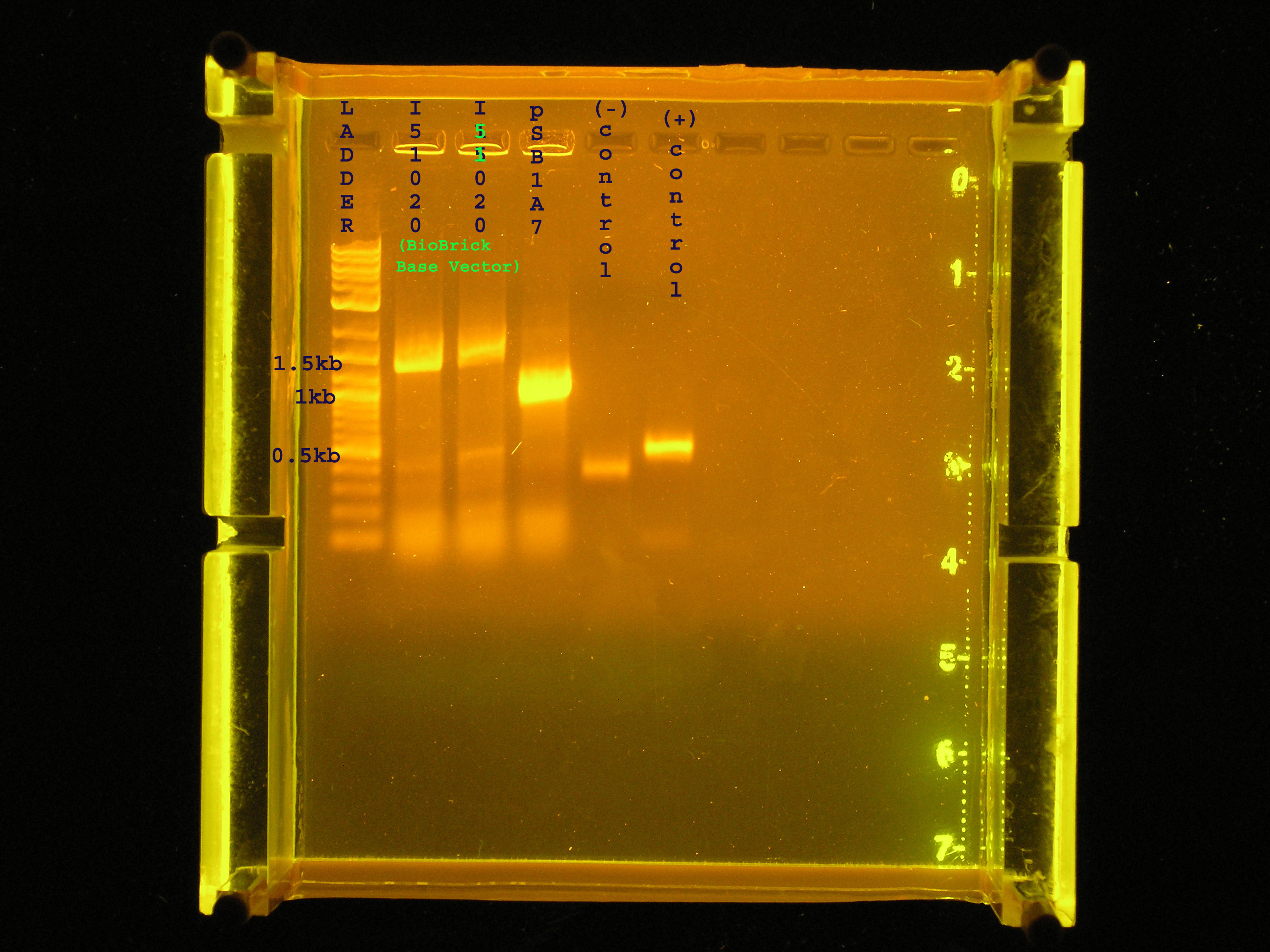
| - | + | [[Image:Colony_PCR_basevector_psb1a7.jpg|right|thumb|300px|Colony PCR verification of the BioBrick Base Vector ([http://partsregistry.org/Part:BBa_I51020 BBa_I51020])and [http://partsregistry.org/Part:pSB1A7 pSB1A7].]] | |
:*Verifying the inserts of two I51020 (the base vector) and one colony of pSB1A7 (an insulated vector with high copy #) | :*Verifying the inserts of two I51020 (the base vector) and one colony of pSB1A7 (an insulated vector with high copy #) | ||
| + | |||
| + | <strong>Results</strong> | ||
| + | |||
| + | :*Lanes 2 and 3 (base vector)contain bands that are approximately 1.5kb, while I would expect the base vector to have a band of 2045. Lane 4 (pSB1A7) contains a band at approximately 1.2 kb while I expect the band to be 1.265 kb. | ||
| + | :*The negative control, UPA amplifying water, contains a faint band indicating an algal contamination in my PCR water. | ||
| + | :*The positive control, UPA amplifying algal DNA is at 0.5kb as expected. | ||
| + | |||
| + | <strong> Conclusions</strong> | ||
| + | |||
| + | :*It is unclear whether the base vector has been isolated. The bands are relatively close in size, indicating they are the same material so there is a chance something got botched in the running of this gel. I will plasmid prep this and verify again with PCR and run the whole plasmid to see if it is what I would expect. | ||
| + | |||
| + | :*I need to use different controls that will give me a better indication of whether or not my methods are sufficient. For example, I can use, as a positive control, colonies from the pSB1A3 plate which was verified on [[Team:Hawaii/Notebook/2008-07-21|7-21.]] | ||
| + | |||
===[[Team:Hawaii/PCR Amplification of pRL1383a|PCR amplification of pRL1383a]]=== | ===[[Team:Hawaii/PCR Amplification of pRL1383a|PCR amplification of pRL1383a]]=== | ||
:<strong> Margaret</strong> | :<strong> Margaret</strong> | ||
Revision as of 02:48, 2 August 2008
| Projects | Events | Resources | ||
|---|---|---|---|---|
| Sponsors | Experiments | Milestones | Protocols | |
| Notebook (t) | Meetings (t) |
Things we did today
Wetlab work
Transformation
- Margaret
- Transformation of I14032 into Db3.1
- Transformation of pRL1383a parts: R0010 plasmid::rep, pSB1A3::oriV, pSB1AS3::oriT
- Check transformation on 7/29/08: oriT had a colony--> restreaked it, and I14032 had 1 colony-->restreaked
Colony PCR
- Margaret
- Verifying the inserts of two I51020 (the base vector) and one colony of pSB1A7 (an insulated vector with high copy #)
Results
- Lanes 2 and 3 (base vector)contain bands that are approximately 1.5kb, while I would expect the base vector to have a band of 2045. Lane 4 (pSB1A7) contains a band at approximately 1.2 kb while I expect the band to be 1.265 kb.
- The negative control, UPA amplifying water, contains a faint band indicating an algal contamination in my PCR water.
- The positive control, UPA amplifying algal DNA is at 0.5kb as expected.
Conclusions
- It is unclear whether the base vector has been isolated. The bands are relatively close in size, indicating they are the same material so there is a chance something got botched in the running of this gel. I will plasmid prep this and verify again with PCR and run the whole plasmid to see if it is what I would expect.
- I need to use different controls that will give me a better indication of whether or not my methods are sufficient. For example, I can use, as a positive control, colonies from the pSB1A3 plate which was verified on 7-21.
PCR amplification of pRL1383a
- Margaret
- The omega region from pSMC121 and the rep/mob region are giving primer dimers and mis-amplification respectively. I am trying a gradient PCR from 49°C to 55°C to see if this will fix the problem.
PCR for BB-pRL1383a construction/verification
- Grace
- PCR of BB-pRL1383a (from plasmid prep) using VF2 & VR_B0034 primers to verify B0034 insert and MCS replacement
- PCR of B0034 with HindIII and BamHI primers in case we need to redo BB-pRL1383a construction
- Maintained everything on ice this time
- Used 0.5μl plasmid in PCR reaction
- Annealing at 55C
- Extension for 30 sec. at 72C
- Gel returned 3 bands + huge smear for B0034. Smear = lots of correct product. What are the three other bands (~120bp, ~170bp, ~200bp)?
- Gel gave two bands for BB-pRL1383a (~170bp, ~300bp)
- Will redo BB-pRL1383a construction using ccdB gene as insert/MCS
Construction of pRL1383a
- Margaret
- Gel purification of R0010::aadA and B0014::mob.
- The bands were really faint. I can either purify and hope the constructs are there, or I can re-do these parts and combine these fractions.(???)
Plasmid preps
- Grace
- Resuspended plasmid preps in 30 μl TE and checked concentrations on nanodrop spectrometer
- nir+B0034 = 13.6 ng/μl
- GFP+B0024 = 9 ng/μl
- GFPf+B0024= 7.1 ng/μl
- BB-pRL1383a = 166.3 ng/μl
- Restriction digested nir+B0034 plasmid prep to verify insert (GFP and GFPf constructs were verified last week)
- Digested w/ EcoRI and SpeI so we can extract from gel if insert is correct
- Incubated at 37C for 2 hours
- Band way too big 1000+bp
- Need to redo
GFP(f) + tt constructs
- Grace
- Restriction digested GFP and GFPf w/ EcoRI and SpeI in 50 μl reactions for 3 hours
- Restriction digested B0024 (tt) with XbaI then EcoRI for 3 hours total
- Ran on an EtBr stained 3.8% agarose gel alongside GFP+B0024 ligation from 7/22 to see if ligation was successful (and the problem was with the colony we picked)
- GFPf+tt did not show a band ~800bp
- GFP ligation was okay the first time around (we picked a bad colony to plasmid prep?)
Reconstruction of assemblies
- Grace & Krystle
- PCR reaction to retrieve ccdB gene and BioBrick MCS from pSB1A3.
- Ran on gel to verify PCR product.
- RE digested overnight in 20 μl reactions (10μl DNA):
- pRL1383a with HindIII in NEBuffer 2
- GFP with EcoRI and SpeI in NEBuffer EcoRI + BSA
- GFP fusion with EcoRI and SpeI in NEBuffer EcoRI + BSA
- nir with EcoRI and SpeI in NEBuffer EcoRI + BSA
- rbs (B0030) with XbaI in NEBuffer 2 + BSA
- tt (B0024) with XbaI in NEBuffer 2 + BSA
Drylab Work
Primer Design
- Margaret
- I want to design primers for some replication proteins + origin from pSB2K3
- Submitted the primers for the P1 lytic replication proteins/origin of replication from pSB2K3 to Norman for review.
Discussion
restriction digests
- the BamHI and HindIII enzymes we have are several years old, we need to remember to run undigested product as well to test if it is really cutting what we want to cut.
Quote of the Day
History is the only laboratory we have in which to test the consequences of thought. - Étienne Gilson
[http://manoa.hawaii.edu/  ][http://manoa.hawaii.edu/ovcrge/
][http://manoa.hawaii.edu/ovcrge/  ][http://www.ctahr.hawaii.edu
][http://www.ctahr.hawaii.edu  ]
]
 "
"