Team:Hawaii/Notebook/2008-08-13
From 2008.igem.org
(Difference between revisions)
(added margaret's list) |
(→Transformation) |
||
| (3 intermediate revisions not shown) | |||
| Line 41: | Line 41: | ||
:::* GFPf likely J33207 due to mix up -- see below | :::* GFPf likely J33207 due to mix up -- see below | ||
| - | + | ||
===Plasmid Prep=== | ===Plasmid Prep=== | ||
<strong> Margaret </strong> | <strong> Margaret </strong> | ||
| Line 51: | Line 51: | ||
:*I corrected my math from yesterday's ligation and re-did the ones that did not ligate correctly | :*I corrected my math from yesterday's ligation and re-did the ones that did not ligate correctly | ||
| + | |||
| + | <strong> Krystle </strong> | ||
| + | |||
| + | :*Ligated using quick ligation buffer and quick ligase: | ||
| + | ::*gfpf+B0015(digested by MR) | ||
| + | ::*gfpf+B0015(digested by GK) | ||
| + | ::*gfp+B0015(digested by MR) | ||
| + | ::*B0015(digested by MR) <-- as a control | ||
===Transformation=== | ===Transformation=== | ||
| Line 57: | Line 65: | ||
:*omega ligation from yesterday | :*omega ligation from yesterday | ||
:*rep+B0030, P1+B0015, aadA(pRL1383a) +B0030 from today's ligation | :*rep+B0030, P1+B0015, aadA(pRL1383a) +B0030 from today's ligation | ||
| + | |||
| + | <strong>Krystle</strong> | ||
| + | :*Transformed using DB3.1 | ||
| + | :*Ligation products from today (see above) | ||
| + | ::Notes: Initial incubation on ice only 3 minutes, final incubation with SOC only 1 hour | ||
===Gel From Yesterday's Colony PCR=== | ===Gel From Yesterday's Colony PCR=== | ||
Latest revision as of 02:44, 15 August 2008
| Projects | Events | Resources | ||
|---|---|---|---|---|
| Sponsors | Experiments | Milestones | Protocols | |
| Notebook (t) | Meetings (t) |
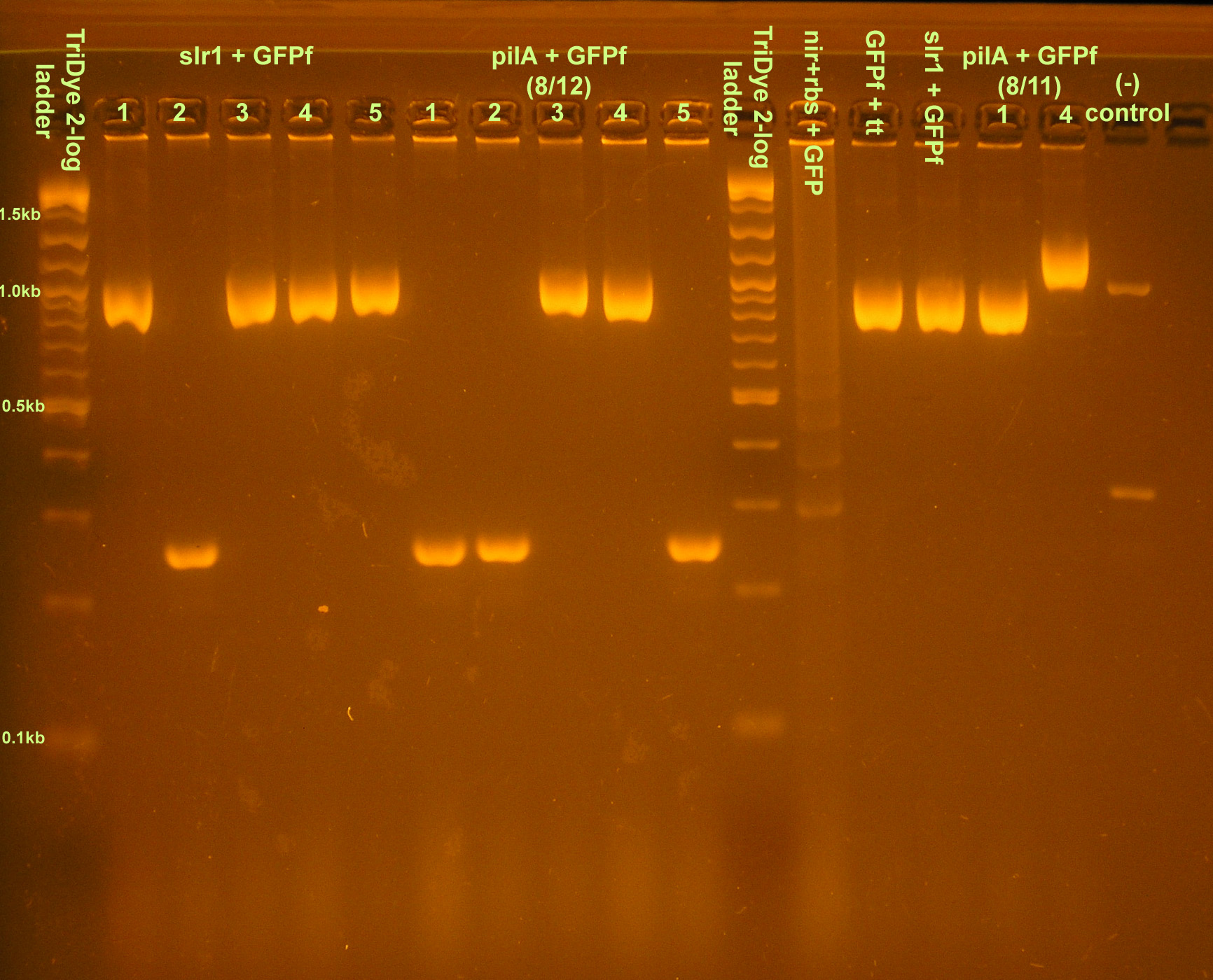
Things we did today
Wetlab work
Verification of Transformants
- Grace
| Construct | Colony forming units |
|---|---|
| slr1 + GFPf | 69 + 1 cluster of colonies |
| pilA + GFPf | 17 |
| nir+rbs + GFP | 0 |
| nir+rbs + slr1 | 0 |
| nir+rbs + pilA | 1 |
| plac+rbs + GFP | 0 |
| plac+rbs + slr1 | 0 |
| plac+rbs + pilA | 0 |
- Colony PCR of transformants and restreaked colonies from yesterday
- Ran on 2.5% agarose gel at 95V for 100 min.
- Restreaked:
- slr1+GFPf colony 5
- pilA+GFPf colony 4
- GFPf likely J33207 due to mix up -- see below
Plasmid Prep
Margaret
- OriV1-4 and aadA(BB) 4 **the only correct transformants from yesterday's ligation/transformation
Ligation
Margaret
- I corrected my math from yesterday's ligation and re-did the ones that did not ligate correctly
Krystle
- Ligated using quick ligation buffer and quick ligase:
- gfpf+B0015(digested by MR)
- gfpf+B0015(digested by GK)
- gfp+B0015(digested by MR)
- B0015(digested by MR) <-- as a control
Transformation
Margaret
- omega ligation from yesterday
- rep+B0030, P1+B0015, aadA(pRL1383a) +B0030 from today's ligation
Krystle
- Transformed using DB3.1
- Ligation products from today (see above)
- Notes: Initial incubation on ice only 3 minutes, final incubation with SOC only 1 hour
Gel From Yesterday's Colony PCR
Margaret
Drylab Work
Sequencing
- Grace
- Checked sequencing results returned from CORE Hawaii
- Confirmed:
- B0015
- B0030
- B0034 (one bp off)
- nir
- slr
- slr1 = slr2 huh?
- Other:
- nir+rbs
- No rbs present; only nir
- Huh? We ligated nir INTO rbs
- I14032+rbs
- No plac present; only rbs
- BB-pRL1383a
- E0040 (GFP) went in successfully, not J33207
- WTF? Our plasmid preps were/are mislabeled
- Redo ligation to get blue/white screen?
- GFP+tt (reverse only)
- Low quality read; GFP is not present (segment too small)
- GFPf+tt (reverse only)
- lacZ fragment present; no tt
- Huh? We ligated the part INTO tt
- GFPf+tt and J33207+tt samples switched?
- J33207+tt
- Part did not go in; tt only
Discussion
Quote of the Day
History is the only laboratory we have in which to test the consequences of thought. - Étienne Gilson
[http://manoa.hawaii.edu/  ][http://manoa.hawaii.edu/ovcrge/
][http://manoa.hawaii.edu/ovcrge/  ][http://www.ctahr.hawaii.edu
][http://www.ctahr.hawaii.edu  ]
]
 "
"