Team:Hawaii/PCR Amplification of pRL1383a
From 2008.igem.org
(added 7/23) |
(added quantities of DNA) |
||
| Line 62: | Line 62: | ||
*running apparatus | *running apparatus | ||
*gel running conditions: 95V for ~1 hour | *gel running conditions: 95V for ~1 hour | ||
| + | |||
| + | <strong> PCR Clean-up using Qiagen Minelute Kit </strong> | ||
| + | |||
| + | #add 5 volumes of PB to 1 volume of PCR product | ||
| + | #place sample in minelute spin column | ||
| + | #centrifuge 1 minute at 13k rpm | ||
| + | #discard flow-through | ||
| + | #add 750uL PE buffer to wash; centrifuge 1 minute 13k rpm | ||
| + | #discard flow-through; centrifuge 1 min 13k rpm | ||
| + | #place column in clean 1.5 mL tube | ||
| + | #add 10ul TE or water to center of column, wait 1 minute, centrifuge 1 min | ||
=== Results === | === Results === | ||
| Line 397: | Line 408: | ||
!width="100"|Completed PCR | !width="100"|Completed PCR | ||
!width="100"|Cleaned? | !width="100"|Cleaned? | ||
| + | !width="100"|Quantity(gel,nanodrop) | ||
!width="100"|Description | !width="100"|Description | ||
| + | !width="100"|Any product remaining? | ||
|- | |- | ||
|OriV | |OriV | ||
|yes | |yes | ||
|yes | |yes | ||
| + | |50ng/ul, 170.9 | ||
|origin of vegetative replication | |origin of vegetative replication | ||
| + | |no | ||
|- | |- | ||
|rep region | |rep region | ||
|yes | |yes | ||
|yes | |yes | ||
| + | |30ng/uL, 238.8ng/ul | ||
|proteins necessary for autonomous replication | |proteins necessary for autonomous replication | ||
| + | |no | ||
|- | |- | ||
|mob region | |mob region | ||
|yes | |yes | ||
|yes | |yes | ||
| + | |30ng/ul, 211.9 ng/ul | ||
|mobilization proteins & origin of transfer | |mobilization proteins & origin of transfer | ||
| + | |no | ||
|- | |- | ||
|rep/mob region | |rep/mob region | ||
| - | |||
|no | |no | ||
| + | |no | ||
| + | |0,0 | ||
|mob and rep proteins/necessary for mobilization by Ti plasmid | |mob and rep proteins/necessary for mobilization by Ti plasmid | ||
| + | |n/a | ||
|- | |- | ||
|omega interposon | |omega interposon | ||
|no, primer dimers | |no, primer dimers | ||
|no | |no | ||
| + | |0,0 | ||
|insertional mutagenesis & Sm, Sp resistance | |insertional mutagenesis & Sm, Sp resistance | ||
| + | |n/a | ||
|- | |- | ||
|aadA gene | |aadA gene | ||
|yes | |yes | ||
|yes | |yes | ||
| + | |52ng/ul, 48.1ng | ||
|Sm, Sp resistance | |Sm, Sp resistance | ||
| + | |yes | ||
|} | |} | ||
{{Team:Hawaii/Footer}} | {{Team:Hawaii/Footer}} | ||
Revision as of 02:15, 26 July 2008
| Projects | Events | Resources | ||
|---|---|---|---|---|
| Sponsors | Experiments | Milestones | Protocols | |
| Notebook (t) | Meetings (t) |
Contents |
PCR Amplification of pRL1383a
- Several regions of pRL1383a will be amplified with BioBrick based primers. These components will be used later in the construction of a pRL1383a BioBrick based vector. These parts include the aadA region of the omega interposon, the origin of vegetative replication (oriV), the mobilization proteins, and the replication proteins.
Methods
Materials Per Reaction Added in order:
- 3.5uL nanopure water
- 1uL 10uM forward/reverse primers
- 1uL pRL1383a as template (1:10 dilution of large-scale plasmid prep).
- 5uL Taq polymerase: Accusure(hot start, 68°C) and Red Taq
Note: the taq and water can be combined and aliquoted together as long as reaction is kept at 4°C.
- cold blocks
- pcr reaction tubes, with tops
- pipette and tips
| Duration of Time | T° 5 cycles | T° 30 cycles | Purpose |
|---|---|---|---|
| 10 minutes | 95°C | 95°C | heat activate taq |
| 30 seconds | 95°C | 95°C | denaturation |
| 30 seconds | 48.1°C, 53.4°C, 57.9°C | 59°C, 60.6°C, 61.9°C | annealing |
| 5 minutes | 68°C | 68°C | extension (1.5 minutes per kb, go with longest) |
| 10 minutes | 68°C | 68°C | finishing the extension |
| infinity | 4°C | 4°C | (until you remove product) |
Materials for the gel
- 1% agarose gel
- 10ug/mL Ethidium Bromide
- running apparatus
- gel running conditions: 95V for ~1 hour
PCR Clean-up using Qiagen Minelute Kit
- add 5 volumes of PB to 1 volume of PCR product
- place sample in minelute spin column
- centrifuge 1 minute at 13k rpm
- discard flow-through
- add 750uL PE buffer to wash; centrifuge 1 minute 13k rpm
- discard flow-through; centrifuge 1 min 13k rpm
- place column in clean 1.5 mL tube
- add 10ul TE or water to center of column, wait 1 minute, centrifuge 1 min
Results
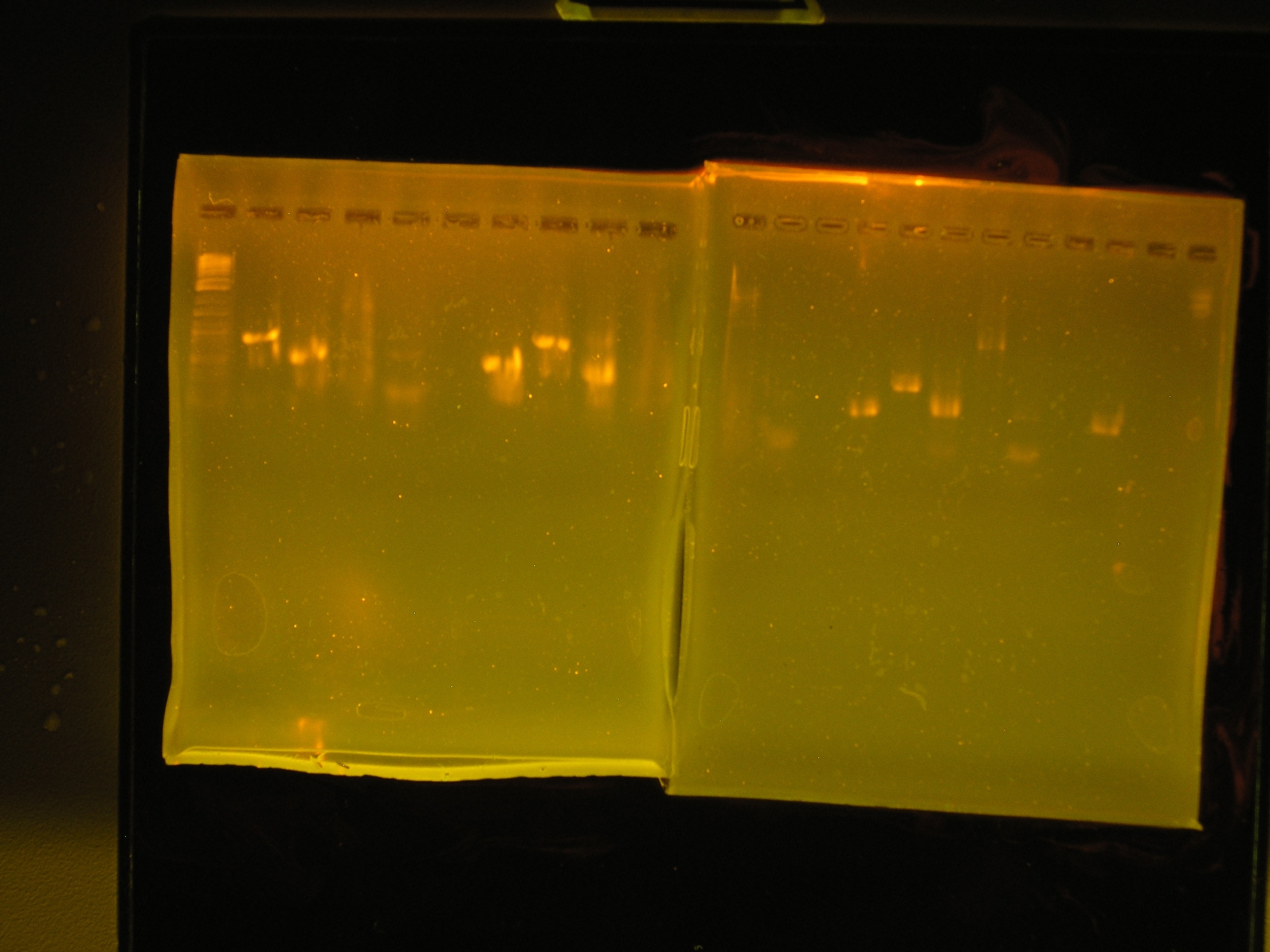
| Lane | Contents | Description | Predicted annealingT°C,5 cycles | Predicted annealing T°C,30 cycles |
|---|---|---|---|---|
| 1 | Tri-dye 10kb ladder | ok | ||
| 2 | omega (aadA)#1 | (+), ~1kb | 54.4,50.4 | 60.7,69.5 |
| 3 | oriV #1 | (+), ~0.5kb | 51.3, 53 | 60.9, 71.2 |
| 4 | mob #1 | smear | 53.5, 48 | 62, 69.8 |
| 5 | rep #1 | smear, low kb | 53.9, 57.2 | 59.1, 73.5 |
| 6 | (-)control | all clear | ||
| 7 | (+) control | bright band, ~0.5kb | ||
| 8 | omega #2 | (+), ~1kb | 54.4,50.4 | 60.7,69.5 |
| 9 | oriV #2 | (+) ~0.5kb | 51.3, 53 | 60.9, 71.2 |
| 10 | mob #2 | nothing, slight smear | 53.5, 48 | 62, 69.8 |
| Lane | Contents | Description | Predicted annealing T°C,5 cycles | Predicted annealing T°C,30 cycles |
|---|---|---|---|---|
| 1 | Tri-dye 10kb ladder | no ladder | ||
| 2 | rep#2 | light smear | 53.9, 57.2 | 59.1, 73.5 |
| 3 | (-)control | all clear | ||
| 4 | (+)control | bright band ?0.5kb | ||
| 5 | omega #3 | bright band, ?1kb | 54.4,50.4 | 60.7,69.5 |
| 6 | oriV #3 | bright band, 0.5kb? | 51.3, 53 | 60.9, 71.2 |
| 7 | mob | light band, some smear, 3kb? | 53.5, 48 | 62, 69.8 |
| 8 | rep #3 | band in low kb | 53.9, 57.2 | 59.1, 73.5 |
| 9 | (-) control | all clear | ||
| 10 | (+) control | bright band, 0.5kb? |
PCR Clean-up
MinElute PCR Purification Kit Protocol from Qiagen using a microcentrifuge
This protocol is designed to purify double-stranded DNA fragments from PCR reactions resulting in high end-concentrations of DNA (see page 12). Fragments ranging from 70 bp to 4 kb are purified from primers, nucleotides, polymerases, and salts using MinElute spin columns in a microcentrifuge.
- Mix PB 5:1 PCR reaction.
- Place a MinElute column in a provided 2 ml collection tube.
- Apply ALL of sample to the MinElute column; centrifuge (13,000rpm)1 min.
- Discard flow-through. Place the MinElute column back into the same tube.
- Add 750 μl Buffer PE to the MinElute column; centrifuge (13,000rpm) 1 min.
- Discard flow-through and place the MinElute column back in the same tube.
Centrifuge the column for an additional 1 min at maximum speed.
- Place the MinElute column in a clean 1.5 ml microcentrifuge tube.
- Add 10 μl 1x TE to the center of the membrane, let the column stand for 1 min; centrifuge 1 min.
Discussion
- The gels are very warped. I did not let them dry long enough before I poured the running buffer,so some products did not run correctly therefore this experiment will be repeated on Monday when the necessary reagents are delivered.
- I mixed up the temperature gradient!!! I need to be more careful about labeling my reactions.
- The rep protein did not come out at all and the mob region only came out at one temperature, so these must be PCRed again to determine the correct annealing temperature.
Follow-up Experiments
7/2/08
- A second attempt was made to PCR the rep region. An annealing temperature of 53.4° was used for the first 5 cycles and annealing temperature 60.6 was used for the subsequent 30 cycles.
- Results: There are 2 faint bands in the low kb region. This is probably primers.
- Discussion: The rep region did not amplify under these conditions. There were several suggestions made by the professors that I will try this weekend.
7/4/08
- Materials:
- 5uL Red Taq (per reaction)
- 3.5uL nanopure water (per reaction)
- 1uL 10uM f/r rep region primers (per reaction)
- 1uL of either 1:100, 1:500, 1:1000 dilution pRL1383a as template (1:10 dilution of large-scale plasmid prep)
- 5uL accusure (for one reaction to test the dilution with this enzyme)
| Duration of Time | T° 5 cycles | T° 30 cycles | Purpose |
|---|---|---|---|
| 10 minutes | 95°C | 95°C | heat activate taq |
| 30 seconds | 95°C | 95°C | denaturation |
| 30 seconds | 54 | 59 | annealing |
| 5 minutes | 70°C (red taq),68°C (accusure) | 68°C (red taq),68°C (accusure) | extension (1.5 minutes per kb, go with longest) |
| 10 minutes | 70°C (red taq),68°C (accusure) | 68°C (red taq),68°C (accusure) | finishing the extension |
| infinity | 4°C | 4°C | (until you remove product) |
Results
| Lane | Contents | Description |
|---|---|---|
| 1 | Tri-dye 10kb ladder | faint, not well defined |
| 2 | rep 1:100 dilution | 3.3kb, brighter than ladder, very thick band, smear (too much DNA?) |
| 3 | rep 1:500 dilution | 3.3kb, brighter than ladder, very thick band, smear (too much DNA?) |
| 4 | rep 1:1000 dilution | 3.3kb, brighter than ladder, very thick band, smear (too much DNA?) |
| 5 | (-)control | very faint band in low kb region (maybe contamination from other well |
| 6 | (+) control | faint band in 0.5 kb region |
| 7 | 1:500 dilution rep region amplified by accusure | 0.5kb band and band in low kb region, faint |
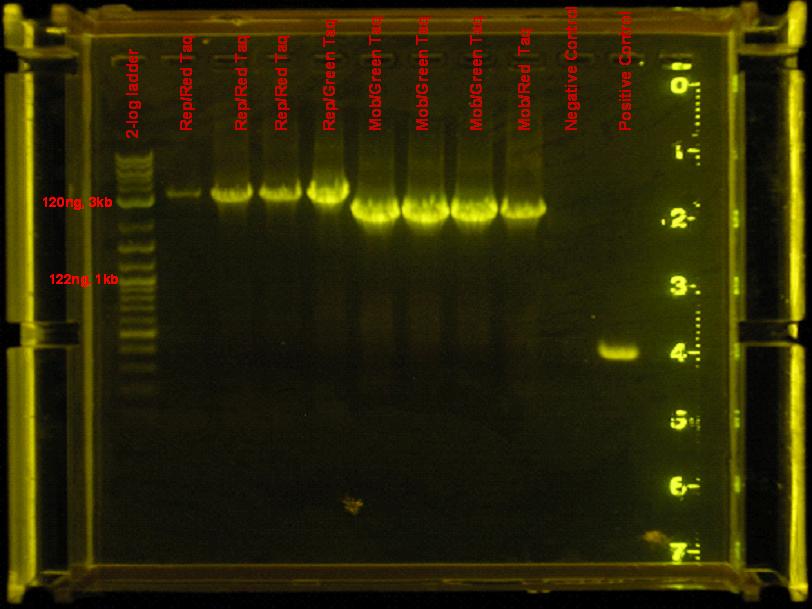
7/7/08
- rep and mob regions did not amplify with accusure, so they were amplified using red/green taq. Same conditions from 7/4 were used.
results
- mob and rep regions were amplified by both red and green taq. Use green in the future, it is cheaper.
Discussion
rep region appears to be amplified by red taq efficiently. Need to use much less DNA. Find out what dilution to use. Accusure + some reaction stabilizing reagent might be used to get this reaction going, but as of right now, this region is not being amplified by this enzyme. This temperature seems to have worked well also.
7/18/08
- PCR amplification of oriV, omega interposon, aadA gene, & rep/mob all using green taq.
- PCR conditions: 2min@94°C, 30sec@95°C, 30sec@50.4°C, 5min30sec@72°C(1 minute per kilobase), infinity@4°C
7/19/08
- PCR amplification of omega interposon and rep/mob regions using 3 different types of taq: File:Velocity taq.pdf, green taq and accuzyme with the hopes that one will amplify these products. I also changed the annealing temperature to 48°C in order to accomodate the primer with the lowest annealing temperature.
Methods For all reactions, I heated up the PCR block to 94C before adding my sample. I did this by running another method which only involves 94C for infinity.
- Accuzyme:
- Reaction conditions: 3.5ul H2, 1ul Primers, 0.5ul template, 5ul taq
- Running conditions: 95C 10min, 95C 30sec, 48C 30sec, 68C 10 min, 68C 10min, 4C infinity.
- Green Taq:
- Reaction conditions: 3.5ul H2, 1ul Primers, 0.5ul template, 5ul taq
- Running conditions: 95C 10min, 95C 30sec, 48C 30sec, 68C 10 min, 68C 10min, 4C infinity.
- Velocity DNA Polymerase:
- Reaction conditions: 2ul 5X Buffer, 0.1ul dNTPs, 5.65ul H2O, 1ul Primers, 0.5ul template, 0.25ul taq(0.5U)
- Running conditions: 98C 2min, 98C 30sec, 48C 30sec, 72C 2min34sec, 72C 10min, 4C infinity.
Results
- As seen in the very small picture, the only amplification product of this reaction is of the positive control amplified by green taq.
- The omega interposon template (pSMC121) was run on a gel alongside the products of the colony PCR from 7_21. There was no band meaning that there was no template for this PCR reaction.
Discussion
- Need to get in touch with Dr. Callahan and get more template. Also, just try again with green taq only... problem solve.
7/23
- PCR amplification of the omega interposon region of pSMC121 and the mob/rep region of pRL1383a using green taq.
- Reaction conditions: same as above
- Running conditions: same as above for green taq.
Results
- The omega interposon amplification yielded only primer dimers. The mob/rep region is a smear, there is a lot of mis-priming. The positive and negative controls yield a bright band and no band respectively.
Discussion
- the omega interposon primers are likely to form dimers because there are 4 overlapping nucleotides at the 5' and 3' ends with a Gibbs free energy of -6.61 kcal/mole. I can alleviate this problem by finding a higher annealing temperature than 48°C. The annealing temperature is 49.5°, so I can use a gradient to find a good temperature. I will try from 48deg&;C to 55°C.
- The above stated method for solving the primer dimer problem can also be a possible solution for the mis-priming of the mob/rep region.
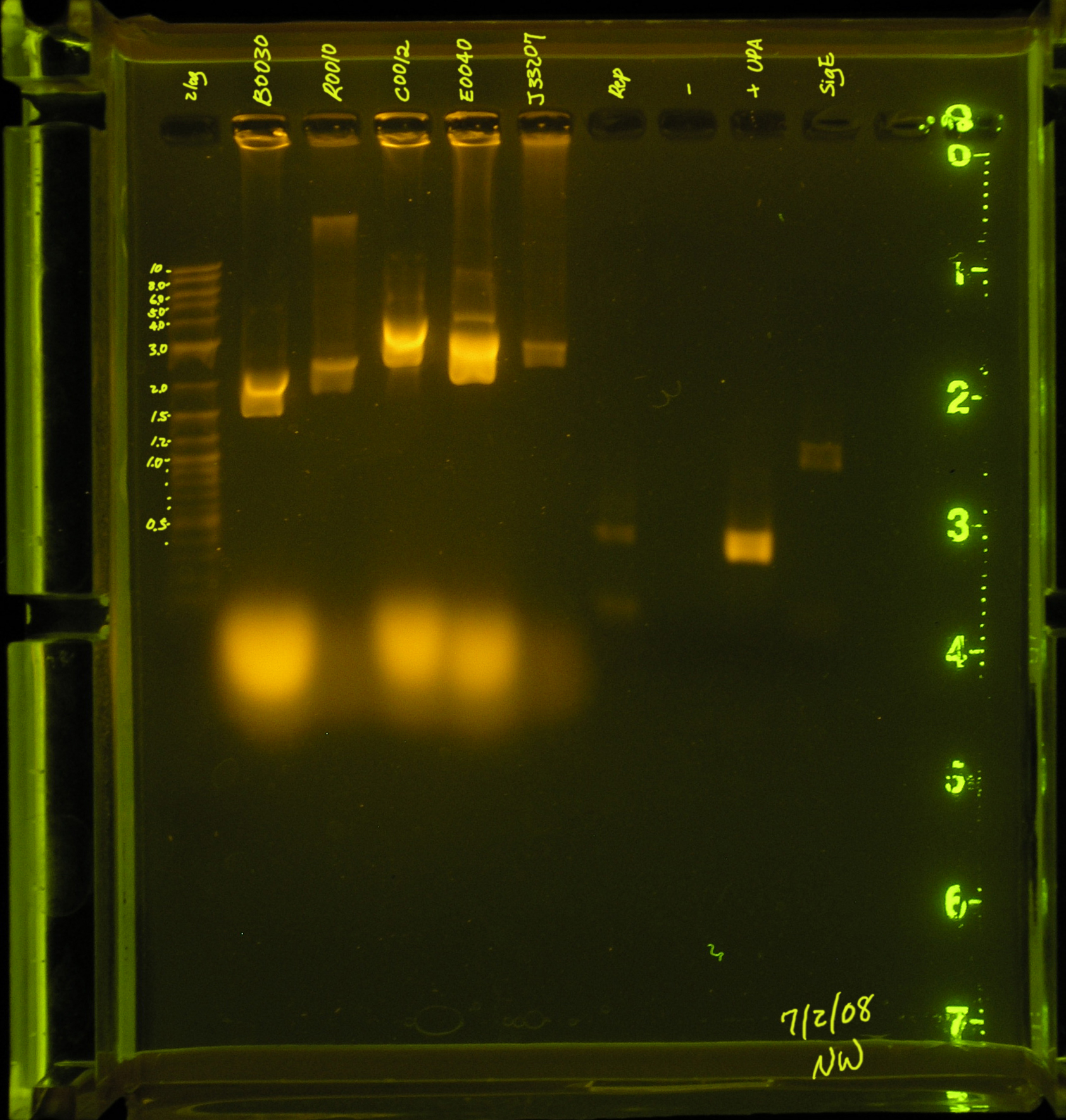
| Name | Completed PCR | Cleaned? | Quantity(gel,nanodrop) | Description | Any product remaining? |
|---|---|---|---|---|---|
| OriV | yes | yes | 50ng/ul, 170.9 | origin of vegetative replication | no |
| rep region | yes | yes | 30ng/uL, 238.8ng/ul | proteins necessary for autonomous replication | no |
| mob region | yes | yes | 30ng/ul, 211.9 ng/ul | mobilization proteins & origin of transfer | no |
| rep/mob region | no | no | 0,0 | mob and rep proteins/necessary for mobilization by Ti plasmid | n/a |
| omega interposon | no, primer dimers | no | 0,0 | insertional mutagenesis & Sm, Sp resistance | n/a |
| aadA gene | yes | yes | 52ng/ul, 48.1ng | Sm, Sp resistance | yes |
[http://manoa.hawaii.edu/  ][http://manoa.hawaii.edu/ovcrge/
][http://manoa.hawaii.edu/ovcrge/  ][http://www.ctahr.hawaii.edu
][http://www.ctahr.hawaii.edu  ]
]
 "
"