Team:Hawaii/Project
From 2008.igem.org
| Projects | Events | Resources | ||
|---|---|---|---|---|
| Sponsors | Experiments | Milestones | Protocols | |
| Notebook (t) | Meetings (t) |
Overall Project
A BioBrick toolkit for cyanobacteria
- We aim to extend the current BioBrick registry to a greater range of organisms, including cyanobacteria. Cyanobacteria are studied for their ability to produce useful compounds, including biofuels and biopolymers. These "little green factories" require only salts, light, water, and carbon dioxide for photoautotrophic growth. A cyanobacterial "toolkit" would enhance our ability to utilize this system.
- We designed:
- 1) mobilizable broad-host range BioBrick vectors derived from RSF1010,
- 2) a cassette for protein secretion from Synechocystis sp. PCC 6803, and
- 3) a nitrate-inducible cyanobacterial promoter BioBrick.
- 1) mobilizable broad-host range BioBrick vectors derived from RSF1010,
- Our toolkit was designed for conjugative gene transfer from Escherichia coli to Synechocystis to achieve the controlled production and recovery or bioproducts, demonstrable by induced secretion of green fluorescent protein. Though our parts were targeted for work in cyanobacteria, they may be compatible with other Gram-negative systems including Agrobacterium, which is capable of plant transformation.
Project Details
Part A: Mobilizable Broad-Host-Range Plasmid
- RSF1010 is a naturally occurring broad-host-range plasmid capable of conjugative transfer and stable replication due to the presence of mob genes with an associated origin of transfer (oriT) and rep genes with an associated origin of vegetative replication (oriV), respectively. We aim to compartmentalize a derivative of the RSF1010 plasmid, namely pRL1383a, into BioBricks. The resulting BioBricks can be inserted into a BioBrick base vector to create a plasmid that transfers genetic elements via conjugation. (read more...)
Part B: Cyanobacterial protein secretion system
- Photosynthetic cyanobacteria provide the opportunity for autotrophic production of practically any biomolecule. The ability to extract engineered biomolecules would make this bacterium a renewable, nearly self-sustaining "factory"- a potentially valuable tool in bioengineering. For Part B, we will create BioBricks encoding naturally occurring signal peptides that can be combined with a protein coding sequence in order to express the protein of interest extracellularly. (read more...)
Materials, Methods, and Results
Part A: Mobilizable Broad-Host-Range Plasmid
Part B: Expression system for Synechocystis sp. PCC 6803
Step 1: Synthesis and assembly of the nirA promoter and pilA and slr2016 signal sequences
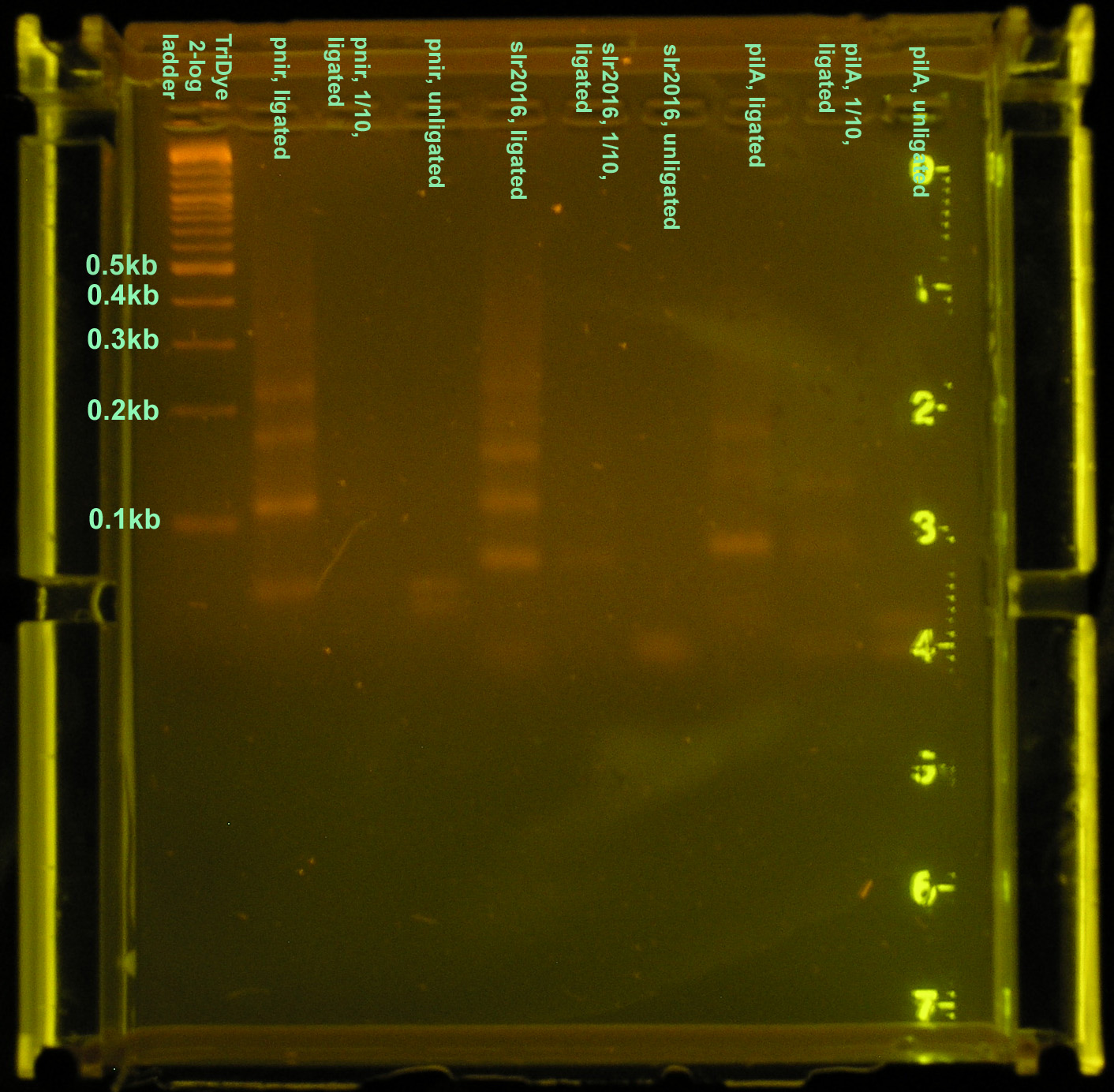
- The nir promoter and the pilA and slr2016 secretion signal sequences were syntheized with the standard Biobrick sites. Oligonucleotide fragments of each were hybridized with its complement and ligated together to form whole, fully functional promoters and signal sequences. Assembly of these new Biobricks were verified by gel electrophoresis and sequencing.
nir promoter
| Oligonucleotide | Sequence | Length |
|---|---|---|
| pnir1_fb.syn.1 | CTAGAGCTAAATGCGTAAACTGCATATGCCTTCGCTGAGTGTAATTTACGTTACA | 55 |
| pnir2_fb.syn.1 | AATTTTAACGAAACGGGAACCCTATATTGATCTCTACTACTAGTAGCGGCCGCTGCA | 57 |
| pnir2_rb.syn.1 | GCGGCCGCTACTAGTAGTAGAGATCAATATAGGGTTCCCGTTTCGTTAAAATTTGTAAC | 59 |
| pnir1_rb.syn.1 | GTAAATTACACTCAGCGAAGGCATATGCAGTTTACGCATTTAGCT | 45 |
Signal sequences
| Oligonucleotide | Sequence | Length |
|---|---|---|
| pilA1_fp.syn.1 | CTAGATGGCTAGTAATTTTAAATTCAAACTCCTCTCTCAAC | 41 |
| pilA2_ff.syn.1 | TCTCCAAAAAACGGGCAGAAGGTGGTACTAGTAGCGGCCGCTGCA | 45 |
| pilA2_rf.syn.1 | GCGGCCGCTACTAGTACCACCTTCTGCCCGTTTTTTGGAGAGTTGAG | 47 |
| pilA1_rp.syn.1 | AGAGGAGTTTGAATTTAAAATTACTAGCCAT | 31 |
| slr2016-1_fp.syn.1 | CTAGATGGCAGCAAAACAACTATGGAAAATTTTCAATC | 38 |
| slr2016-2_ff.syn.1 | CTAGACCGATGAAGGGTGGAACTAGTAGCGGCCGCTGCA | 39 |
| slr2016-2_rf.syn.1 | GCGGCCGCTACTAGTTCCACCCTTCATCG | 29 |
| slr2016-1_rp.syn.1 | GTCTAGGATTGAAAATTTTCCATAGTTGTTTTGCTGCCAT | 40 |
Three different constructs of the full nir promoter and slr2016 and pilA signal sequences were created. The first followed ligation and restriction digest protocols using full concentrations of annealed products. The second was a ligation and restriction digest carried out using 10-1 dilutions of annealed product. It was suggested by Dr. Sean Callahan that if too much DNA was added to the ligation mixture, "DNA hairballs" would form between ssDNA (unannealed) and dsDNA, resulting in smears and multiple bands in the gel. The last method of construction simply placed equal concentrations of annealed products together. In theory, DNA overhangs would anneal appropriately, yielding an unligated but full promoter or signal sequence.
Ligation at full concentrations of annealed product worked best. The strongest bands were observed for this method. Multiple bands were still observed, above and below the expected band length (80-100bp) indicating both unligated DNA fragments and incompletely digested DNA or hairball structures. However, the strongest band for all three constructs corresponded to the desired product. The 10-1 ligations were barely visible on the gel, and multiple bands were still observed, so ligation and restriction digest efficiency was not improved by the addition of less DNA. The last method of annealing but not ligating yielded two bands in the gel corresponding to the two original DNA fragments.
The purified DNA was then ligated to a Biobrick vector, pSB1A2, for subcloning in E. coli. Ligations were carried out using 3:1 molar concentrations of insert to vector.
- Sequencing by the Greenwood Molecular Biology Facility returned the following results:
- nir promoter
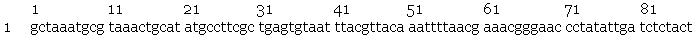
- slr2016 signal sequence

- pilA signal sequence
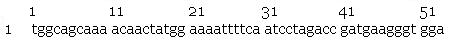
Step 2: Site-directed mutagenesis of GFP (BBa_E0040) into a fusion brick
- GFP, as it currently exists in the Biobrick Registry of Parts, is a protein Biobrick, meaning that it will ligate out of frame with our signal sequence Biobricks. A primer will be designed for site-directed mutagenesis of the GFP start codon to convert BBa_E0040 into a fusion Biobrick.
| Primer | Sequence | Length | G/C content | Tm |
|---|---|---|---|---|
| GFP (BBa_E0040) fusion / foward primer | GCCGCTTCTAGAcgtaaaggag | 22 bp | 54.55% | 60.2 C |
| GFP (BBa_E0040) fusion / reverse primer | cgagtcagtgagcgaggaag | 20 bp | 60% | 59.6 |
Step 3: Conversion of pRL1383a into a Biobrick plasmid
- Part A of our project focuses on converting the RSF1010 based plasmid, pRL1383a into a sophisticated broad-host Biobrick plasmid. While we aim to ultimately express our secretion system in this new plasmid as part of a cyanobacterial expression system, we need a workable shuttle vector between E. coli (where constructs will be made) and PCC6803 (the ultimate host). Converting pRL1383a into a much simpler Biobrick plasmid will fulfill this requirement. Verification regions, transcriptional terminators, and the Biobrick multiple cloning site (MCS) will be isolated from the plasmid containing BBa_J33207 via PCR. PCR primers will also include HindIII and BamHI restriction sites for ligation into pRL1383a. This ligation will replace the original pRL1383a MCS which includes Biobrick and Biobrick compatible restriction sites. The MCS replacement will be verified by restriction digest and plasmid sequencing.
| Primer | Sequence | Length | G/C content | Tm | Notes |
|---|---|---|---|---|---|
| HindIII-VF2BB | cctAAGCTTtgccacctgacgtctaagaa | 29 bp (20 bp) | 48.3% (50.0%) | 65.9 C (58.6 C) | Includes RE extension HindIII site and three 5' nucleotides for efficient cutting. Parentheses indicate primer information w/o RE site and 3 nucleic acids. Based on VF2 primer. |
| BamHI-VRBB | ccaGGATCCattaccgcctttgagtgagc | 29 bp (20 bp) | 55.2% (50.0%) | 67.9 C (58.0 C) | Includes RE extension BamHI site and three 5' nucleotides for efficient cutting. Parentheses indicate primer information w/o RE site and 3 nucleic acids. Based on VR primer. |
Step 4: Device construction
- The synthesized signal peptides and nirA promoter BioBricks will be combined with at least three existing BioBricks to create two (or more) nitrate-regulated protein secretion devices according to the scheme detailed in Figure 1. The resulting devices will be placed in a Synechocystis compatible BioBrick vector derived from the RSF1010 derived plasmid pRL1383a. In the proposed devices, the signal peptides will be situated so they are in-frame with GFP. The translated polypeptide should consist of a N-terminal signal polypeptide leader sequence attached to a fluorescent protein.
Step 5: Testing for protein secretion
- The BioBrick vector can be inserted into Synechocystis sp. PCC6803 by triparental conjugation with E. coli harboring a transmissible plasmid (like RP1) and another E. coli containing our engineered plasmid. Plated Synechocystis sp. PCC6803 colonies successfully transformed wwill exhibit a glowing halo of secreted GFP. Transformed Synechocystis sp. PCC6803 grown in liquid media will result in fluorescent culture media. The efficacy of the signal peptides in transporting GFP into the extracellular media can be measured using a spectrofluorometer.
Controls
- A number of controls will also be constructed in parallel with our device to test device assembly and function.
| Construct | Control | Test |
|---|---|---|
| nirA + rbs (BBa_B0034) + GFP (BBa_E0040) + txn term. (BBa_B1006) | lac promoter (BBa_I14032) + rbs (BBa_B0034) + GFP (BBa_E0040) + txn term. (BBa_B1006) | Inducibility of nirA promoter |
| nirA promoter + rbs (BBa_B0034) + pilA or slr2016 signal sequence + GFP fusion brick + txn term. txn term. (BBa_B1006) | nirA or lac promoter + rbs (BBa_B0034) + GFP (BBa_E0040) + txn term. (BBa_B1006) | Functionality of secretion peptides and GFP fusion |
Results
Part A: Mobilizable Broad-Host-Range Plasmid
Expression system for Synechocystis sp. PCC 6803
Implications and future plans
 "
"