Team:Heidelberg/Notebook/Killing I/Notebook/week5
From 2008.igem.org
(Difference between revisions)
(New page: <html> <link rel='stylesheet' href='http://igem-heidelberg.de/fileadmin/Wiki/Heidelberg2.css' type='text/css' /> <link rel="stylesheet" href="http://igem-heidelberg.de/fileadmin/Wiki/Menu....) |
|||
| Line 120: | Line 120: | ||
</body> | </body> | ||
</html> | </html> | ||
| + | |||
'''Week 5''' | '''Week 5''' | ||
Revision as of 22:14, 25 October 2008


Week 5
Contents |
Monday, 09/01/08
conjugation test
- donor: pUB307, pED374
- recipient: pBAD33
- protocol
- donor: pUB307, pED374
- mix 2ml/ 1,5ml/ 1ml donor culture with 1ml/ 1,5ml/ 2ml recipient culture
- incubate for 1h at 37°C
- plate 0,1ml on amp & cm each and incubate at 37°C for h
first phage cloning strategy
- Nanodrop evaluation
- pBAD Maxiprep 138,1 ng/ul; 1,99
- pBluescript Maxiprep 280 ng/µl; 1,97
- glycerol stock (2 aliquot corresponding to two different colonies) of pUB307(helper plasmid) and pED374 (OriT + MOB)
Digestion of PCR fragments
- GFP digestion 1
74µl purified PCR DNA 10µl NEB1 10µl BSA 3µl KpnI 3µl AgeI
- 2h at 37°, 300rpm
- PCR purification kit with GFP digestion 1
- GFP digestion 2
30 µl DNA from purified GFP digestion 1 5µl NEB3 5µl 3µl ScaI 12µl water
- 2h at 37°, 300rpm
- digestion of GAM (purified PCR DNA)
60µl DNA 12µl water 10µ NEB1 10µl BSA 2µl AgeI 3µl XhoI (75% activity in buffer)
- digestion of OriT (purified PCR DNA)
75µl DNA 1ßµl NEB2 10µl BSA 3µl KpnI (75% activity in buffer) 3µl XbaI
- digestion of CmR (purified PCR DNA)
60µl DNA 27µl water 10µl NEB3 3µl ScaI
- digestion of pBluescript (vector)
50µl DNA 24µl H20 3µl XbaI 3µl XhoI 10µl NEB2 10 µl BSA
- purification of the fragments with PCR purification kit
- ligation of the fragments (18h, 14°C)
GAM: 7µl oriT: 7µl GFP: 7µl CmR: 7µl pBluescript: 6µl 10x ligase buffer: 4µl T4 DNA ligase: 2µl
- new PCR of CmR, GFP, oriT and GAM --> same programm as before
- digestion of I20260 in pSB3K3 (GFP) with ScaI (over night, 14°C)
Tuesday, 09/02/08
conjugation test
- conjugation test worked --> we got many colonies
- new conjugation test to quantify results:
- 2ml donor mixed with 1ml recipient, incubated for 1h at 37°C, plated on AmpR and CmR and incubated for 16h - 2 samples were made
- the following dilutions were made and plated:
- conjugation 1 and 2: plated undiluted, 10^-3, 10^-4, 10^-5
- donor plated: 10^-4, 10^-5, 10^-6
- recipient plated: 10^-4, 10^-5, 10^-6
- evaluation
- titer of donor: 1,84*10^9/ml
- titer of recipient:3,4*10^9/ml
- ratio of donor and recipient: 1,082
- titer of conjugation samples:
- sample 1: 5,93*10^5
- sample 2: 5,4*10^5
- conjugation efficiency (percentage of donor cells that got a plasmid):
- sample 1: 0,0174%
- sample 2: 0,0159%
new phage cloning strategy
- transformation of the ligated vector in Top10/DH5alpha and MG1655 (always 4µl DNA in 100µl cells)
- brought out on amp+cm plate
- Purifcation of the PCR products and the I20260 digest (after heat shock)
- Nanodrop evaluation of the PCR products
- OriT 147,4 ng/µl; 1,91
- GAM 118,3 ng/µl; 1,90
- GFP 184,2 ng/µl; 1,89
- I20260 29,9 ng/µl; 0,94
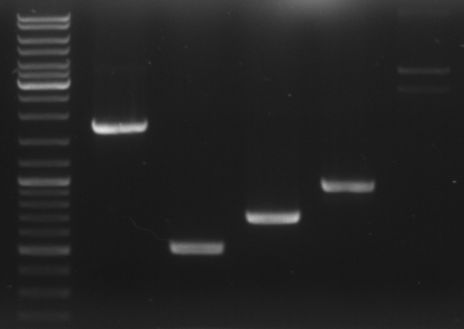
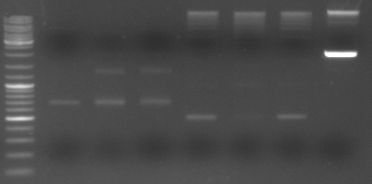
- loading all the PCR fragments (1µl each) on a gel
- lane0: ladder
- lane1: GAM fragment(~1.44kb)
- lane2: oriT (~0.52kb)
- lane3: CmR (~0.66kb)
- lane4: GFP (~0.92kb)
- lane5: I20260 (digested with ScaI --> one single cut --> linearised ~3.7kb)
--> PCR worked --> I20260 seems not to be right
- digestion of I20260 as indicated above
- digestion of lambda DNA with XbaI and XhoI to get more of the big vector fragment (as indicated before)
- digestion of GAM, OriT, CmR, GFP for cloning, protocols are listed at the first try above
- digestion of pBluescript as testvector for the Derbyshire OriT
30µl DNA from Maxiprep 5µl water 5µl NEB2 5µl BSA 2µl XbaI 3µl KpnI
- Purification and heat inactivation for all digestions
- ligation of CmR in I20260
- transformation on Kan+Cm plate
- ligation of OriT and pBluescript over night
6µl insert 4µl vector 2µl Buffer (10x) 1µl Ligase 7µl nuclease free water
Wednesday, 09/03/08
conjugation test
- conjugation test: we still have a lot of colonies so we need to dilute it further. But still we could reproduce our conjugation results
new phage cloning strategy
- Transformation of OriT and PBluescript ligation
- plated out on Xgal + IPTG + Amp plate
- we ordered our cI at Geneart
- pick colony of CmR::I20260 transformation for liquid culture (miniprep)
- start making new competent cells (TOP10)
- make DH5alpha AUII (prey)MgSO4-Stock
- Titering the DH5alpha AUII (prey)MgSO4-Stock on LB/Amp plates
- Titer: 8.2*10^8
- infection test: Lambda cI 857 (in vitro packaging)-DH5alpha AUII(prey)
- result: positive. The infection rate could be a little lower than WT
- parameter measurement for m (decay-rate of Lambda)
- test Lambda: Lambda cI 857 of in vitro packaging
- test buffer: M9 and Standard I-Maltose
- test time: 0h-8h
- test temperature: 42°C
- indicator bacteria: DH5alpha WT
- protocol see parameter measurement protocol
- parameter measurement for b (adsorption-rate of Lambda)
- test Lambda: Lambda cI 857 of in vitro pachaging
- test bacteria: DH5alpha AUII (prey)
- test time: 0min-18min
- test temperature: 37°C
- indicator bacteria: DH5alpha WT
- protocol see parameter measurement protocol
- test failed
Thursday, 09/04/08
conjugation test
- for quantitative measurement of conjugation: get neubauer chamber (for cell counting)
- conjugation test: test different conjugation times and donor/recipient ratios:
- stop conjugation after 60, 75, 90 and 120 min
- ratios donor/recipient 1:2 and 2:1
- plate conjugation diluted 1:10^3 and 1:10^4
- alltogether 16 plates
- cell titer is measured via neubaur chamber
new phage cloning strategy
- inoculation of pBluscript liquid culture from glycerol stock for new maxiprep
- Test the Derbyshire OriT
- liquid culture of OriT and pBluescript ligation (4 cultures, 10ml LB, Amp)
- Ligation of GAM, OriT, CmR, GFP in one vector
- miniprep of CmR::I20260
- results of measurement of DNA concentration from miniprep of GFP containing CmR:
- 77 ng/µl, 260/280: 1,62
- 8,3 ng/µl, 260/280: 2,47
- 12,1 ng/µl, 260/280: 2,57
- 16,5 ng/µl, 260/280: 1,67
- 13,4 ng/µl, 260/280: 1,6
- 8,3 ng/µl, 260/280: 2,4
- 8,1 ng/µl, 260/280: 2,64
- 8,1 ng/µl, 260/280: 1,79
- 12,2 ng/µl, 260/280: 2,39
- 14,0 ng/µl, 260/280: 2,36
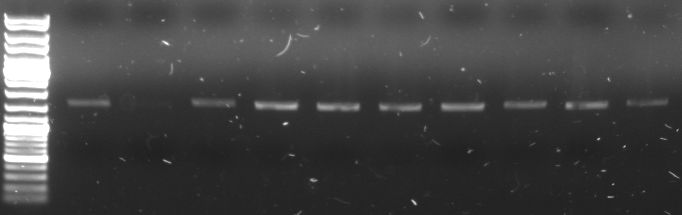
- I20260::CmR plasmid DNA used as a template for PCR with GFP Primer to get GFP cassette with CmR in it (Phusion)
- gel:
- lane0: DNA ladder mix
- lane1-10: GFP+CmR (1,58 kb)
- all PCR products (without the one from lane2) were combined for PCR purification --> elution in 30µl
- digestion of GFP::CmR with KpnI/AgeI
- purify digested fragment
- digestion of pBluescript with XbaI, XhoI (Vector)
- ligate purified CmR::GFP with purified GAM/oriT/pBluescript
- testtransformation on Amp+CmR
Friday, 09/05/08
conjugation test
- conjugation test did not work at all - no colonies
new phage cloning strategy
- we got colonies on the control plate (pBAD33 on Amp and Cm)! so we have to test pBAD33 again on Amp and Cm. If pBAD33 is amp resistant, all conjugative tests done so far would be useless.
- plated pBAD33 on Amp
- ligation of OriT, GAM, CmR, GFP in vector
- Maxiprep of pBluescript
- miniprep of oriT in pBluescript
- digestion of OriT in pBluescript, digestion with BglI to check the plasmid
- nanodrop results of preps:
- pBluescript:: OriT
- 158,1 ng/µl, 260/280: 1,89
- 156,2 ng/µl, 260/280: 1,88
- 98,8 ng/µl, 260/280: 1,94
- 168,0 ng/µl, 260/280: 1,91
- 164,6 ng/µl, 260/280: 1,89
- 125,9 ng/µl, 260/280: 1,85
- 154,4 ng/µl, 260/280: 1,84
- 127,2 ng/µl, 260/280: 1,98
- Maxiprep pBluescript: 2793,5 ng/µl, 260/280: 1,92
- pBluescript:: OriT
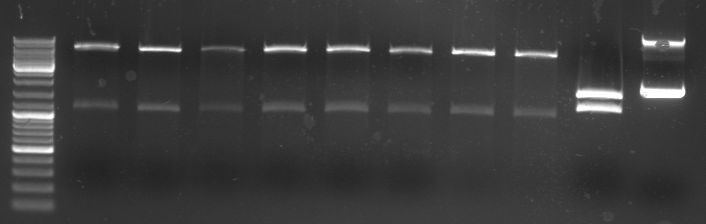
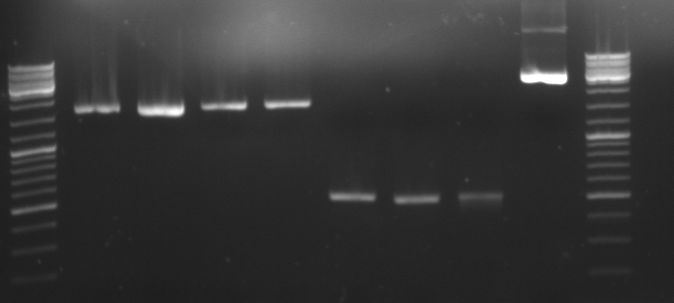
- Gels of digestion and ligation:
- lane 0: DNA ladder mix
- lane 1-8: oriT in pBluescript diegested with BglI (2100/1270bp)
- lane 9: new pBluescript digested with BglI 1 (1700/1270)
- lane 10: old pBluescript undigested (2960bp)
- lane 0: DNA ladder mix
- lane 1-6: ligation of 4 parts (GFP::CmR) in pBluescript (not yet transformed)
- lane1-3: ligation at 4°C for 16h
- lane4-6: ligation at 14°C for 16h
- lane 7: old pBluescript undigested (2960bp)
- conclusions
- the pBluescript::OriT is probably wrong
- the old Maxiprep of pBluescript is probably wrong
- but OriT might also be wrong
- we can't be sure if our ligations just didn't work because a part was wrong and should check the fragments we are working with
New proceeding:
- just to check
- transformation of oriT in pBluescript together with pUB307 (helper plasmid)
- transformation of ligation of 4 parts in pBluescript
- but at the same time:
- next week inoculation of liquid culture of new donor bacteria --> glycerol stocks --> conjugation tests
- New digestions of the new pBluescript Maxis
- with XbaI-XhoI
- with XbaI-AgeI
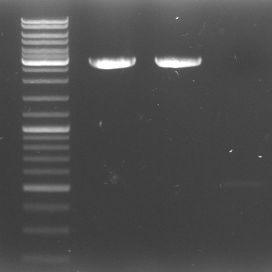
- To check our fragments: The-what-the-heck-have-we-actually-got-gel
- lane 0: DNA ladder mix
- lane 1: GFP::CmR Verdau (1.58kb)
- lane 2: GFP::CmR PCR Purificated 4.9 (1.58kb)
- lane 3: GAM 2.9 (1.44)
- lane 4: GAM geschnitten (1.44kb)
- lane 5: OriT 1 (0,52kb)
- lane 6: OriT 2 (0,52kb)
- lane 7: OriT digested purificated 4.9 (0.52kb)
- lane 8: pBluescript neu unverdaut 1:10 Verdünnung (2.96kb)
- lane 9: DNA ladder
- lane0: DNA ladder mix
- lane1: pBluescript XbaI/KpnI
- lane2: pBluescript XbaI/XhoI
- lane3: oriT aus pBluescript (0.52kb)
 "
"