Team:Heidelberg/Notebook/Killing II/1stweek
From 2008.igem.org
(Difference between revisions)
(→Cloning) |
(→Friday 08/08/2008) |
||
| (7 intermediate revisions not shown) | |||
| Line 481: | Line 481: | ||
- incubate overnight at 37 °C | - incubate overnight at 37 °C | ||
*Planings for Receiver - Colicin cloning: | *Planings for Receiver - Colicin cloning: | ||
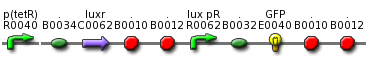
| - | ** use part BBa_T9002 and substitute GFP with colicin genes <br> [[Image: | + | ** use part BBa_T9002 and substitute GFP with colicin genes <br> [[Image: BBa T9002.png| 400 px| center]] |
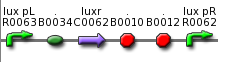
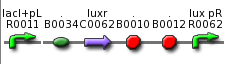
| - | ** use part BBa_F2621 or BBa_F2622 insert colicin cassette behind Lux P<sub>R</sub> (MCS) <br> [[Image: | + | ** use part BBa_F2621 or BBa_F2622 insert colicin cassette behind Lux P<sub>R</sub> (MCS) <br> [[Image: BBa F2621.png| 280 px| center]] <br> [[Image: BBa F2622.png| 280 px| center]] |
===Sender part=== | ===Sender part=== | ||
| Line 497: | Line 497: | ||
- incubate overnight at 37 °C | - incubate overnight at 37 °C | ||
| + | [[https://2008.igem.org/Team:Heidelberg/Notebook/Killing_II/1stweek back]] | ||
==Tuesday 08/05/2008== | ==Tuesday 08/05/2008== | ||
| Line 524: | Line 525: | ||
*Inoculation of liquid cultures for glycerol stocks of amplifier, sender and receiver+GFP parts: For each part we picked 6 colonies and incubated them on LB:antibiotics (1000:1) media over night | *Inoculation of liquid cultures for glycerol stocks of amplifier, sender and receiver+GFP parts: For each part we picked 6 colonies and incubated them on LB:antibiotics (1000:1) media over night | ||
| + | [[https://2008.igem.org/Team:Heidelberg/Notebook/Killing_II/1stweek back]] | ||
==Wednesday 08/06/2008== | ==Wednesday 08/06/2008== | ||
| Line 572: | Line 574: | ||
4 °C constant | 4 °C constant | ||
| + | [[https://2008.igem.org/Team:Heidelberg/Notebook/Killing_II/1stweek back]] | ||
==Thursday 08/07/2008== | ==Thursday 08/07/2008== | ||
| Line 587: | Line 590: | ||
*New plan: If we do not receive the LuxI sender part from the registry, we amplify the LuxI gene from ''Vibrio fischeri'' genome. For that we have to design primers with restriction sites for pBAD MCS: | *New plan: If we do not receive the LuxI sender part from the registry, we amplify the LuxI gene from ''Vibrio fischeri'' genome. For that we have to design primers with restriction sites for pBAD MCS: | ||
| - | + | [[https://2008.igem.org/Team:Heidelberg/Notebook/Killing_II/1stweek back]] | |
==Friday 08/08/2008== | ==Friday 08/08/2008== | ||
*Theoretical Work... | *Theoretical Work... | ||
| + | |||
| + | [[https://2008.igem.org/Team:Heidelberg/Notebook/Killing_II/1stweek back]] | ||
[[Team:Heidelberg/Notebook/Killing_II/2ndweek | go to 2<sup>nd</sup> week]] | [[Team:Heidelberg/Notebook/Killing_II/2ndweek | go to 2<sup>nd</sup> week]] | ||
Latest revision as of 21:19, 28 October 2008


1st week
Contents |
Monday 08/04/2008
Colicin Receiver
Cloning
- Transformation of miniPrep DNA of Receiver+GFP part (BBa_T9002) for Glycerolstocks in TOP10 cells: 5 µl per 100 µl E. coli TOP 10 competent cells
- start thawing 100 µl competent E. coli TOP 10 cells on crushed ice - add 5 µl ligation - incubate on ice for 20 minutes - heat shock at 42 °C for 45 sec - incubate 2 min on ice - add 400 µl preheated LB media - incubate at 37 °C for 1 h, 500 rpm - Plate 200 µl on preheated LB-Amp plate - incubate overnight at 37 °C
- Planings for Receiver - Colicin cloning:
- use part BBa_T9002 and substitute GFP with colicin genes
- use part BBa_F2621 or BBa_F2622 insert colicin cassette behind Lux PR (MCS)
- use part BBa_T9002 and substitute GFP with colicin genes
Sender part
Cloning
- Transformation of sender (BBa_F1610) and amplifier part (BBa_I15030) (Transformationprotocol Chris): 5 µl per 100 µl E. coli TOP 10 competent cells
- start thawing 100 µl competent E. coli TOP 10 cells on crushed ice - add 5 µl ligation - incubate on ice for 20 minutes - heat shock at 42 °C for 45 sec - incubate 2 min on ice - add 400 µl preheated LB media - incubate at 37 °C for 1 h, 500 rpm - Plate 200 µl on preheated LB-Amp plate - incubate overnight at 37 °C
[back]
Tuesday 08/05/2008
Colicin Receiver
Cloning
- Transformation results:
- many colonies on receiver plates
- Transformation BBa_F2621 and BBa_F2622 in TOP 10 cells (Transformationprotocol Chris): 5 µl per 100 µl E. coli TOP 10 competent cells
- start thawing 100 µl competent E. coli TOP 10 cells on crushed ice - add 5 µl ligation - incubate on ice for 20 minutes - heat shock at 42 °C for 45 sec - incubate 2 min on ice - add 400 µl preheated LB media - incubate at 37 °C for 1 h, 500 rpm - Plate 200 µl on preheated LB-Amp plate - incubate overnight
Activity tests
- Inoculation of liquid cultures with TOP10 and receiver+GFP transformed TOP10 cells for a quantitative measurement of the GFP expression induced by AI-1.
Sender part
Cloning
- Transformation results:
- small colonies on sender and amplifier plates
- Inoculation of liquid cultures for glycerol stocks of amplifier, sender and receiver+GFP parts: For each part we picked 6 colonies and incubated them on LB:antibiotics (1000:1) media over night
[back]
Wednesday 08/06/2008
Colicin Receiver
Cloning
- Mail to help@registry.com to order BBa_F1610, BBa_I15030, BBa_F2621 & BBa_F2622 because several transformation attempts did not work.
- Transformation results (08/05/2008):
- BBa_F2621 and BBa_F2622 in TOP 10 cells were not grown
- Glycerol Stocks of Receiver+GFP TOP10 cells (BBa_T9002)
- PCR for amplification of BBa_T9002, BBa_F1610, BBa_I15030
25.0 µl Phusion MasterMix 2.5 µl Primer fw 2.5 µl Primer rv 17.0 µl H2O 3.0 µl DNA
program 98 °C 2 min 98 °C 30 sec | 48 °C 30 sec | 25 cycles 72 °C 1 min 30 sec | 72 °C 5 min 4 °C constant
Activity tests
- Quantitative measurement of GFP Expression induced by AI-1: pipetting scheme
Sender part
Cloning
- Mail to help@registry.com to order BBa_F1610, BBa_I15030, BBa_F2621 & BBa_F2622 because several transformation attempts did not work.
- New plan: If we do not receive the LuxI sender part from the registry, we amplify the LuxI gene from Vibrio fischeri genome. For that we have to design primers with restriction sites for pBAD MCS:
- Liquid cultures from previous day (08/05/2008):
- sender (BBa_F1610) transformed cells were not grown
- amplifier (BBa_I15030) transformed cells were not grown
- PCR for amplification of BBa_T9002, BBa_F1610, BBa_I15030
25.0 µl Phusion MasterMix 2.5 µl Primer fw 2.5 µl Primer rv 17.0 µl H2O 3.0 µl DNA
program 98 °C 2 min 98 °C 30 sec | 48 °C 30 sec | 25 cycles 72 °C 1 min 30 sec | 72 °C 5 min 4 °C constant
[back]
Thursday 08/07/2008
Colicin Receiver
Cloning
- Mail to help@registry.com to order BBa_F1610, BBa_I15030, BBa_F2621 & BBa_F2622 because several transformation attempts did not work.
- Gelextraction of Receiver PCR (06 Aug 2008)
Activity tests
- Due to the high OD and fluorescence values we use M9 media for our next AHL measurements.
Sender part
Cloning
- Mail to help@registry.com to order BBa_F1610, BBa_I15030, BBa_F2621 & BBa_F2622 because several transformation attempts did not work.
- New plan: If we do not receive the LuxI sender part from the registry, we amplify the LuxI gene from Vibrio fischeri genome. For that we have to design primers with restriction sites for pBAD MCS:
[back]
Friday 08/08/2008
- Theoretical Work...
[back]
 "
"