Team:Heidelberg/Project
From 2008.igem.org
Annastoeckl (Talk | contribs) (→Project Details) |
|||
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
<html> | <html> | ||
| - | <link rel='stylesheet' href='http://igem-heidelberg.de/fileadmin/Wiki/ | + | <link rel='stylesheet' href='http://igem-heidelberg.de/fileadmin/Wiki/Heidelberg2.css' type='text/css' /> |
| - | + | <link rel="stylesheet" type="text/css" href="http://igem-heidelberg.de/fileadmin/Wiki/Menu.css" /> | |
| + | <link rel="stylesheet" type="text/css" href="http://igem-heidelberg.de/fileadmin/Wiki/MenuHD.css" /> | ||
<div id="header"><img src="https://static.igem.org/mediawiki/2008/7/78/TeamHeidelbergHeader.jpg" alt="Team Heidelberg" /></div> | <div id="header"><img src="https://static.igem.org/mediawiki/2008/7/78/TeamHeidelbergHeader.jpg" alt="Team Heidelberg" /></div> | ||
| - | <div id="navi"> | + | <div class='MenuBar' id="navi"> |
| - | <ul> | + | <ul> |
| - | <li><span><a href="https://2008.igem.org/Team:Heidelberg">Home</a>< | + | <li> |
| - | <li><span><a href="https://2008.igem.org/Team:Heidelberg/Team">Team</a></span></li> | + | <span><a href="https://2008.igem.org/Team:Heidelberg">Home</a><span> |
| - | <li><span><a href="https://2008.igem.org/Team:Heidelberg/Project">Project</a></span></li> | + | </li> |
| - | <li><span><a href="https://2008.igem.org/Team:Heidelberg/Parts">Parts</a></span></li> | + | <li> |
| - | <li><span><a href="https://2008.igem.org/Team:Heidelberg/Modeling">Modeling</a></span></li> | + | <span><a href="https://2008.igem.org/Team:Heidelberg/Team">Team</a></span> |
| - | <li><span><a href="https://2008.igem.org/Team:Heidelberg/Notebook">Notebook</a></span></li> | + | </li> |
| - | <li><span><a href="https://2008.igem.org/Team:Heidelberg/ | + | <li> |
| - | </ul> | + | <span><a href="https://2008.igem.org/Team:Heidelberg/Project">Project</a></span> |
| - | <br style="clear: both;" /> | + | <ul class="DropDownMenu" id="MB1-DDM1"> |
| - | <img src="https://static.igem.org/mediawiki/2008/e/ef/Navi_bg.gif" alt="" /> | + | <li><a href="https://2008.igem.org/Team:Heidelberg/Project/Sensing"><span><span>Sensing</span></span></a></li> |
| + | <li><a href="https://2008.igem.org/Team:Heidelberg/Project/Killing_I"><span><span>Killing I</span></span></a></li> | ||
| + | <li><a href="https://2008.igem.org/Team:Heidelberg/Project/Killing_II"><span><span>Killing II</span></span></a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | |||
| + | <li> | ||
| + | <a href="https://2008.igem.org/Team:Heidelberg/Parts">Parts</a> | ||
| + | <ul class="DropDownMenu" id="MB1-DDM2"> | ||
| + | <li><a href="https://2008.igem.org/Team:Heidelberg/Parts"><span><span>Submitted Parts</span></span></a></li> | ||
| + | <li><a href="https://2008.igem.org/Team:Heidelberg/Parts/Characterization"><span><span>Characterization</span></span></a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | <li> | ||
| + | <a>Modeling</a> | ||
| + | <ul class="DropDownMenu" id="MB1-DDM3"> | ||
| + | <li><a href="https://2008.igem.org/Team:Heidelberg/Modeling/Chemotaxis"><span><span>Chemotaxis - Killing</span></span></a></li> | ||
| + | <li><a href="https://2008.igem.org/Team:Heidelberg/Modeling/Phage"><span><span>Phage Dynamics - model</span></span></a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | <li> | ||
| + | <a>Notebook</a> | ||
| + | <ul class="DropDownMenu" id="MB1-DDM5"> | ||
| + | <li><a href="https://2008.igem.org/Team:Heidelberg/Notebook/Sensing_Group/Notebook"><span><span>Sensing Group ></span></span></a> | ||
| + | <ul class="SideMenu" id="MB1-DDM2-SM1"> | ||
| + | <li><a href="https://2008.igem.org/Team:Heidelberg/Notebook/Sensing_Group/Cloning"><span>Cloning strategy</span></a></li> | ||
| + | <li><a href="https://2008.igem.org/Team:Heidelberg/Notebook/Sensing_Group/Notebook"><span>Notebook</span></a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | <li><a href="https://2008.igem.org/Team:Heidelberg/Notebook/Killing_I/Notebook"><span><span>Killing I ></span></span></a> | ||
| + | <ul class="SideMenu" id="MB1-DDM2-SM2"> | ||
| + | <li><a href="https://2008.igem.org/Team:Heidelberg/Notebook/Killing_I/Cloning"><span>Cloning strategy</span></a></li> | ||
| + | <li><a href="https://2008.igem.org/Team:Heidelberg/Notebook/Killing_I/Notebook"><span>Notebook</span></a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | <li><a href="https://2008.igem.org/Team:Heidelberg/Notebook/Killing_II/Notebook"><span><span>Killing II ></span></span></a> | ||
| + | <ul class="SideMenu" id="MB1-DDM2-SM3"> | ||
| + | <li><a href="https://2008.igem.org/Team:Heidelberg/Notebook/Killing_II/Cloning"><span>Cloning strategy</span></a></li> | ||
| + | <li><a href="https://2008.igem.org/Team:Heidelberg/Notebook/Killing_II/Notebook"><span>Notebook</span></a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | <li><a href="https://2008.igem.org/Team:Heidelberg/Notebook/material"><span><span>Material & Methods</span></span></a> | ||
| + | <li><a href="https://2008.igem.org/Team:Heidelberg/Notebook/team_meetings"><span><span>Team Meetings</span></span></a> | ||
| + | <li><a href="https://2008.igem.org/Team:Heidelberg/Notebook/seminar"><span><span>Seminar on Synthetic Biology</span></span></a> | ||
| + | </ul> | ||
| + | </li> | ||
| + | <li style="width: 160px"> | ||
| + | <a>Human Practice</a> | ||
| + | <ul class="DropDownMenu" id="MB1-DDM4"> | ||
| + | <li><a href="https://2008.igem.org/Team:Heidelberg/Project/General_information"><span><span>General Project Introduction</span></span></a></li> | ||
| + | <li><a href="https://2008.igem.org/Team:Heidelberg/Project/Science_Communication"><span><span>Scientific Communication</span></span></a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | |||
| + | <li> | ||
| + | <a href="https://2008.igem.org/Team:Heidelberg/Sponsors">Sponsors</a> | ||
| + | </li> | ||
| + | </ul> | ||
| + | <br style="clear: both;" /> | ||
| + | <img src="https://static.igem.org/mediawiki/2008/e/ef/Navi_bg.gif" alt="" / > | ||
</div> | </div> | ||
Revision as of 15:06, 22 October 2008

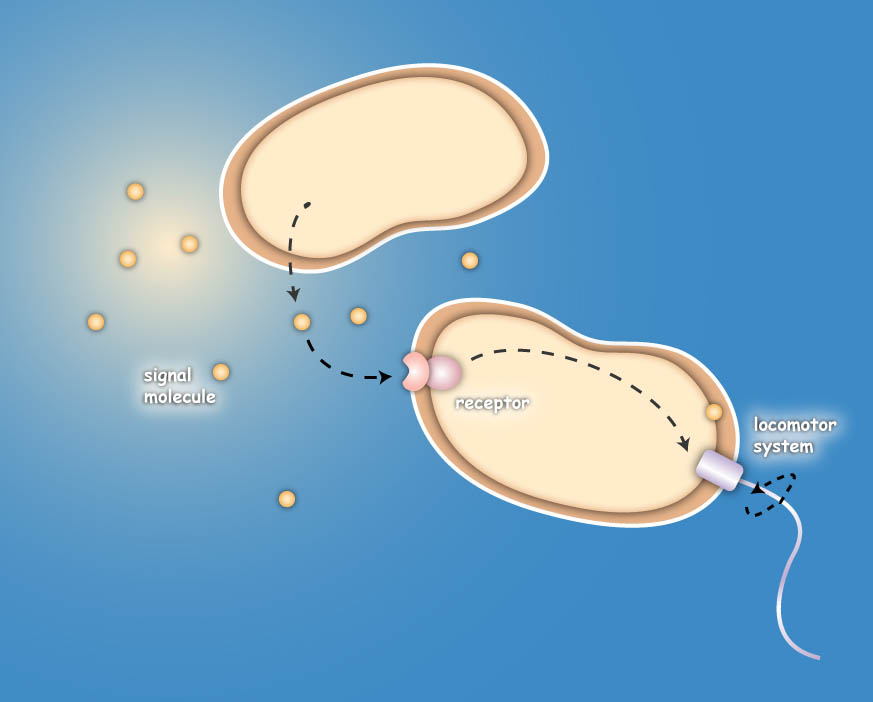
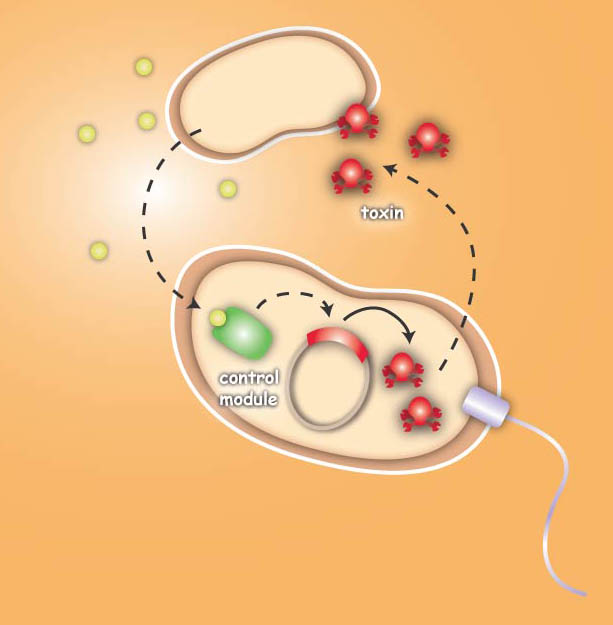
Overall projectEcolicence to kill: Engineering E.coli for targeting pathogenic microorganisms Pathogenic microorganisms represent a major challenge to both medicine and industry. Microbial communities known as biofilms prove to be particularly resistant to conventional therapies and demonstrate the need for alternative methods to fight bacterial growth. The formation of biofilms, as well as other processes such as virulence, antibiotic production, and sporulation, are regulated by communication circuits that depend on small signalling molecules called autoinducers. Our aim is to exploit this communication mechanism by engineering synthetic bacteria that are able to target potentially harmful autoinducer-secreting species and kill them. Working with E. coli as a model system, we plan to engineer separate “killer” and “prey” cells, and divide our project into two complementary modules. The “sensing module” comprises the modification of E. coli’s chemotaxis system to make killer cells move towards prey cells. This requires the engineering of a “sensing module” which propagates a prey-specific stimulus to the downstream signalling cascades, prompting the cells to swim towards the gradient of such a stimulus. The directed swimming stops only in the region where the stimulus is maximal, i.e. in the vicinity of the prey. The “killing module” ensures that once in the vicinity of the prey, a bacteriocidal mechanism is activated. Computer models will show how both modules behave in concert and probe the efficiency of the system in defined spatial environments. Future directions, which are beyond the scope of this project, include the modification of the sensory and killing modules, adjusting the range of targets to real pathogens, but also other organisms, specific cell types or even cancer cells. Project DetailsSensingThe team utilizes the natural sensing and movement system of E. coli bacteria, chemotaxis, by redesigning the receptors of the killer strain to sense a molecular cocktail secreted by the prey strain. After arriving at the prey, an additional gene module is activated which then leads to the death of the pathogen or the cancer cell. The Heidelberg team follows two different strategies to achieve the killing. The first strategy focuses on bacterial toxins, which are normally produced by certain bacterial strains to kill other bacteria. For the test system the team uses genetic information of bacterial toxin production. This information will be introduced into the killer-cells and modified to only become active once the killer strain reaches the prey strain. Activation then leads to toxin production and release, finally resulting in killing of the prey-cell. The second approach for killing uses bacteriophages, which naturally infect E. coli cells. Like many other viruses they kill their prey and replicate into their host. Free phages are then again able to attack other bacteria. In this approach the team specifically takes advantage of this domino effect.
|
Human Practice - Science CommunicationHi, you are new here and hear of synthetic Biology for the first time? Perfect! I am Phips the Phage and will be your guide to this exciting field of biological research and if you like I will explain the background of this project to you. Just follow me…. |
 "
"