Team:Illinois/Project
From 2008.igem.org
| (38 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | + | [[Image:gel.png|950px|center]] | |
| - | + | __NOTOC__ | |
| - | + | <html> | |
| - | + | <head> | |
| - | + | <style> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | .ideasList{ | ||
| + | margin-bottom:30px | ||
| + | } | ||
| + | |||
| + | ul#sub { | ||
| + | margin:0px; | ||
| + | padding:0px; | ||
| + | font-size:14px; | ||
| + | list-style-image:none; | ||
| + | list-style-type:none; | ||
| + | |||
| + | } | ||
| + | |||
| + | #sub li { margin-bottom:20px;} | ||
| + | |||
| + | #sub li a { | ||
| + | color:#3300FF; | ||
| + | text-decoration:underline; | ||
| + | } | ||
| + | |||
| + | #sub li a:hover { | ||
| + | text-decoration:none; | ||
| + | } | ||
| + | |||
| + | .links { | ||
| + | width:100%; | ||
| + | background-color:#FF6103; | ||
| + | margin-bottom:30px; | ||
| + | } | ||
| + | |||
| + | .links tr { | ||
| + | background-color: #FF6103;; | ||
| + | } | ||
| + | |||
| + | .links td:hover { | ||
| + | background-color:#FF6103; | ||
| + | } | ||
| + | |||
| + | |||
| + | .links a{ | ||
| + | text-decoration:none; | ||
| + | font-size:15pt; | ||
| + | font-weight:bold; | ||
| + | font:'helvetica'; | ||
| + | color: rgb(255,255,255); | ||
| + | } | ||
| + | |||
| + | .links a:active { | ||
| + | background-color:#FF6103; | ||
| + | } | ||
| + | |||
| + | .links a:hover { | ||
| + | color: rgb(0,0,128); | ||
| + | } | ||
| + | |||
| + | #navactive { | ||
| + | background-color: rgb(0,0,128); | ||
| + | } | ||
| + | |||
| + | |||
| + | #header { | ||
| + | width: 100%; | ||
| + | text-align: center; | ||
| + | } | ||
| + | |||
| + | h4 { | ||
| + | font-size: 14pt; | ||
| + | font-weight: normal; | ||
| + | } | ||
| + | |||
| + | dt { | ||
| + | font-size: 12pt; | ||
| + | } | ||
| + | |||
| + | #projectnav h4 { | ||
| + | padding: .4ex; | ||
| + | } | ||
| + | |||
| + | #projectnav dt { | ||
| + | font-weight: normal; | ||
| + | margin: 1.5ex 0 0 0; | ||
| + | } | ||
| + | |||
| + | #projectnav h4, #projectnav dt, #projectnav dd { | ||
| + | text-align: center; | ||
| + | text-indent: 0; | ||
| + | margin-left: 0; | ||
| + | } | ||
| + | |||
| + | #hbnav dd { | ||
| + | background-color: #FF6103;; | ||
| + | border-left: dashed #FFF 3px; | ||
| + | border-right: dashed #FFF 3px; | ||
| + | padding: .8ex; | ||
| + | } | ||
| + | |||
| + | #denav dd{ | ||
| + | background-color: #FF6103;; | ||
| + | border-left: dashed #FFF 3px; | ||
| + | border-right: dashed #FFF 3px; | ||
| + | padding: 1ex; | ||
| + | margin-top: 0; | ||
| + | } | ||
| + | |||
| + | #projectnav a { | ||
| + | color: #fff; | ||
| + | } | ||
| + | |||
| + | #pagecontent p { | ||
| + | text-indent: 1.25em; | ||
| + | } | ||
| + | </style> | ||
| + | <! -- END of style sheet --> | ||
| + | </head> | ||
| + | <body> | ||
| + | |||
| + | <!-- Navigation Links Table--> | ||
| + | <table class="links" > | ||
| + | <tr> | ||
| + | <td align="center" ><a class="mainLinks" href="https://2008.igem.org/Team:Illinois" >Home</a> </td> | ||
| + | <td align="center" ><a class="mainLinks" href="https://2008.igem.org/Team:Illinois/Team" >Team</a> </td> | ||
| + | <td align="center"id="navactive" ><a class="mainLinks" href="https://2008.igem.org/Team:Illinois/Project" >Project</a> </td> | ||
| + | <td align="center" ><a class="mainLinks" href="https://2008.igem.org/Team:Illinois/Notebook" >Notebook</a> </td> | ||
| + | <td align="center" ><a class="mainLinks" href="https://2008.igem.org/Team:Illinois/Research_articles" >Research Articles</a> </td> | ||
| + | <td align="center" ><a class="mainLinks" href="https://2008.igem.org/Team:Illinois/Parts" >Parts</a> </td> | ||
| + | <td align="center" ><a class="mainLinks" href="https://2008.igem.org/Team:Illinois/Current_Tasks">Protocols</a> </td> | ||
| + | <td align="center" ><a class="mainLinks" href="https://2008.igem.org/Team:Illinois/Pictures" >Pictures</a> </td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </html> | ||
<br> | <br> | ||
| - | + | == '''Project Abstract''' == | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | |||
| - | + | The unifying motivation behind our research this year is the creation of novel diagnostic tools for medicine. To this end, we are conducting three parallel research projects to create cell-based biosensors. We are currently engineering a bimolecular fluorescence system in which two halves of a fluorescent protein, each fused to an antigenic epitope, will bind to the two sites on an antibody in human serum to cause a detectable fluorescent signal when antibodies against this specific antigen are present. These proteins can be produced in bulk through a bacterial expression system. We are also pursuing similar diagnostic objectives using a eukaryotic system; we are designing strains of yeast able to respond specifically to immunogenic epitopes or antibodies, and activate a fluorometric or enzymatic response accordingly. We are fusing antibodies against immunological targets to cell surface receptors of transcriptional signaling pathways, which would become activated only in the presence of these pathogens. | |
| + | <br><br> | ||
| + | [[Image:Dirty_water.jpg|left|450px]] | ||
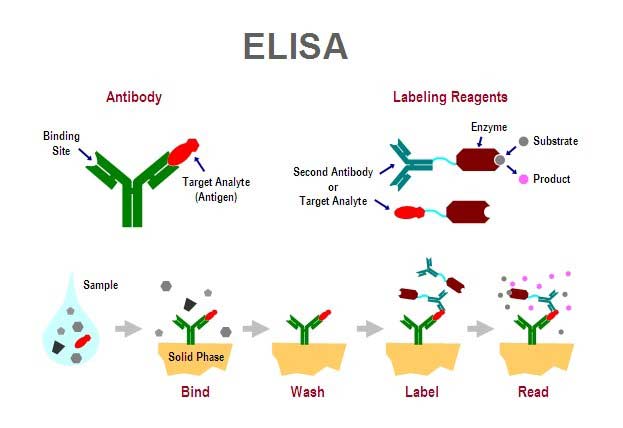
| + | The basic health infrastructure that is enjoyed throughout the developed world is only just begining to emerge in the third world. However, there remains a large need for sanitation and disease detection. Due to the lack of infrastructure in the third world is it more productive to detect and prevent the spread of disease rather than treat the disease once a patient is infected. Unfortunately many detection methods that are routine in the developed world are difficult or impractical throughout the rest of the world. These detection methods, encluding ELISA, DNA sequencing, and protein assays, often require sterile lab conditions, expensive reagents and equipment, and trained personnel. If a simple and robust one-step diagnostic could be developed the implications for public health would be huge. This the the motivation for our projects. | ||
| + | <br><br> | ||
| + | [[Image:elisa.jpg|455px|right|thumb|The mechanism of an Enzyme-Linked Immunorsorbent Assay, a typical method for detecting disease. ]] | ||
| + | Our team is large compared to some, and we can draw on many areas of expertise. It is because of this that we decided to take on the challenge of three separate but related projects. Throughout our brainstorming sessions early in the year we came up with several promising ideas that contained the common thread of medical biosensing. We felt that we gave ourselves the best chance for success by splitting into three subgroups and attacking the same problem from three different angles. Despite being three seperate groups we continued to collaborate closely; each subgroup communicating with and assisting the others. | ||
| + | <br><br> | ||
| + | Throughout the summer and fall we have worked to get our lab up to speed, assemble all the necessary supplies, and build our knowledge base. We have had some successes and some failures but we are committed to continuing on with our projects. We are optimistic that in the future we will have created not only functional, but also useful biosensors for effective pathogen detection in the third world. | ||
== Subprojects == | == Subprojects == | ||
| - | + | === [[Team:Illinois/Antibody_GPCR_Fusion|Antibody GPCR Fusion]] === | |
| + | <html><a href="https://2008.igem.org/Team:Illinois/Antibody_GPCR_Fusion"><img src="https://static.igem.org/mediawiki/2008/a/aa/Gpcr.png" height="165"width="190" align="left"></a></html> | ||
| - | |||
| - | |||
This team will attempt pathogen detection by fusing the appropriate antibody domain to a GPCR receptor and upon binding of the target protein (i.e. cholera toxin), linking the resulting signal cascade to transcription of a reporter, GFP or LacZ. | This team will attempt pathogen detection by fusing the appropriate antibody domain to a GPCR receptor and upon binding of the target protein (i.e. cholera toxin), linking the resulting signal cascade to transcription of a reporter, GFP or LacZ. | ||
| + | <br><br> | ||
| + | <br><br> | ||
=== [[Team:Illinois/Antibody_RTK_Fusion|Antibody Receptor Tyrosine Kinase Fusion]] === | === [[Team:Illinois/Antibody_RTK_Fusion|Antibody Receptor Tyrosine Kinase Fusion]] === | ||
| - | + | <html><a href="https://2008.igem.org/Team:Illinois/Antibody_RTK_Fusion"><img src="https://static.igem.org/mediawiki/2008/9/9f/Rtk.png" height="165"width="190" align="right"></a></html> | |
| + | |||
| + | |||
| + | In this project our plan is to fuse a single-chain antibody to a receptor tyrosine kinase so that the receptor will activate in the presence of cholera toxin. The antibody is specific for the B subunit of the cholera toxin and will cause the receptors to dimerize and activate a signal transduction pathway. | ||
| + | <br><br><br><br> | ||
=== [[Team:Illinois/Bimolecular_Fluorescence_Biosensor|Bimolecular Fluorescence Biosensor]] === | === [[Team:Illinois/Bimolecular_Fluorescence_Biosensor|Bimolecular Fluorescence Biosensor]] === | ||
| - | + | <html><a href="https://2008.igem.org/Team:Illinois/Bimolecular_Fluorescence_Biosensor"><img src="https://static.igem.org/mediawiki/2008/f/f9/Bifc.png" height="165"width="190" align="left"></a></html> | |
| - | + | <br> | |
| + | A soluble biosensor that activates a fluorescent protein upon binding a specific biomolecule | ||
| + | <br><br><br><br><br><br><br> | ||
| - | { | + | {{bottom_template}} |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Latest revision as of 02:12, 30 October 2008
| Home | Team | Project | Notebook | Research Articles | Parts | Protocols | Pictures |
Project Abstract
The unifying motivation behind our research this year is the creation of novel diagnostic tools for medicine. To this end, we are conducting three parallel research projects to create cell-based biosensors. We are currently engineering a bimolecular fluorescence system in which two halves of a fluorescent protein, each fused to an antigenic epitope, will bind to the two sites on an antibody in human serum to cause a detectable fluorescent signal when antibodies against this specific antigen are present. These proteins can be produced in bulk through a bacterial expression system. We are also pursuing similar diagnostic objectives using a eukaryotic system; we are designing strains of yeast able to respond specifically to immunogenic epitopes or antibodies, and activate a fluorometric or enzymatic response accordingly. We are fusing antibodies against immunological targets to cell surface receptors of transcriptional signaling pathways, which would become activated only in the presence of these pathogens.
The basic health infrastructure that is enjoyed throughout the developed world is only just begining to emerge in the third world. However, there remains a large need for sanitation and disease detection. Due to the lack of infrastructure in the third world is it more productive to detect and prevent the spread of disease rather than treat the disease once a patient is infected. Unfortunately many detection methods that are routine in the developed world are difficult or impractical throughout the rest of the world. These detection methods, encluding ELISA, DNA sequencing, and protein assays, often require sterile lab conditions, expensive reagents and equipment, and trained personnel. If a simple and robust one-step diagnostic could be developed the implications for public health would be huge. This the the motivation for our projects.
Our team is large compared to some, and we can draw on many areas of expertise. It is because of this that we decided to take on the challenge of three separate but related projects. Throughout our brainstorming sessions early in the year we came up with several promising ideas that contained the common thread of medical biosensing. We felt that we gave ourselves the best chance for success by splitting into three subgroups and attacking the same problem from three different angles. Despite being three seperate groups we continued to collaborate closely; each subgroup communicating with and assisting the others.
Throughout the summer and fall we have worked to get our lab up to speed, assemble all the necessary supplies, and build our knowledge base. We have had some successes and some failures but we are committed to continuing on with our projects. We are optimistic that in the future we will have created not only functional, but also useful biosensors for effective pathogen detection in the third world.
Subprojects
Antibody GPCR Fusion
This team will attempt pathogen detection by fusing the appropriate antibody domain to a GPCR receptor and upon binding of the target protein (i.e. cholera toxin), linking the resulting signal cascade to transcription of a reporter, GFP or LacZ.
Antibody Receptor Tyrosine Kinase Fusion
In this project our plan is to fuse a single-chain antibody to a receptor tyrosine kinase so that the receptor will activate in the presence of cholera toxin. The antibody is specific for the B subunit of the cholera toxin and will cause the receptors to dimerize and activate a signal transduction pathway.
Bimolecular Fluorescence Biosensor
A soluble biosensor that activates a fluorescent protein upon binding a specific biomolecule
| Home | Team | Project | Notebook | Research Articles | Parts | Protocols | Pictures |
 "
"