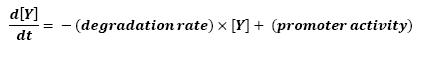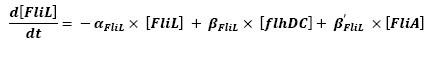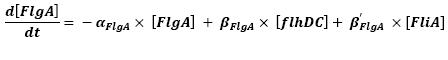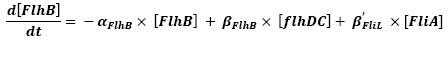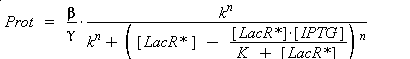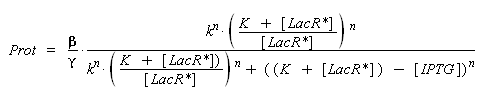An Oscillatory biological Model
Introduction
The goal here is to present the differential equations we used for our system modelization. At each step, we shall describe why we chose this precise model, its drawbacks and possible improvments, the parameters involved and enventually a biologically coherent value.
Bibliography
We whall refer to those three articles :
[1] Shiraz Kalir, Uri Alon. Using quantitative blueprint to reprogram the dynamics of the flagella network. Cell, June 11, 2004, Vol.117, 713-720.
[2]Jordi Garcia-Ojalvo, Michael B. Elowitz, Steven H. Strogratz. Modeling a synthetic multicellular clock : repressilators coupled by quorum sensing. PNAS, July 27, 204, Vol. 101, no. 30.
[3]Nitzan Rosenfeld, Uri Alon. Response delays and the structure of transcription networks. JMB, 2003, 329, 645-654.
[4]Nitzan Rosenfeld, Michael B. Elowitz, Uri Alon. Negative autoregulation speeds the response times of transcription networks. JMB, 2003, 323, 785-793.
Equations
- FlhDC ---> FliL (1)
- FlhDC ---> FlgA (2)
- FlhDC ---> FlhB (3)
- FliA ---> FliL (4)
- FliA ---> FlgA (5)
- FliA ---> FlhB (6)
For all these equations, we found in [1] that in that precise case, the promoter activity the seven class 2 operons, among which FLiL, FlgA, FlhB, may be mathematically described in that way :
promoter activity = βi x [FlhDC] + βi' x [FliA]
where [X] denotes the effective protein-level activity at time.
For each operon, Shiraz Kalir and Uri Alon came up with numerical values of β and β', available in [1].
Furthermore, the protein-level activity can be presented (for a more detailed presentation, see[4]) as
Thus :
Parameters summary
Graph screenshots
Roadmap
If you want to have a look at our roadmap : Roadmap
Bibliography
In order to choose a proper modeling approach for our system, we have decided to list all the chemical reactions we will take into account. Afterwards, we will find the needed parameters reading articles or devising the required experiments.
An overview of the work that has to be done can be found here : Bibliography
Estimation of parameters
If we want to use the promoters used for the formation of the flagella (Description of the project), we will have to clearly defined their dynamics. To do so, a rather huge experimental work will be undertaken, consisting in providing the so-called 'Hill functions' for each promoters.
getting a Hill function from convenient datas
Therefore, we have written a little module which can estimate the parameters of the 'Hill functions', even with some noise and few data available.
Some details and the corresponding code can be found here : Programs.
The method we have employed is just based on a least-square optimization. Then, it could be generic enough for many applications and we would be glad to share the code if you feel it could be usefull.
getting convenient datas
Thus, we need experimental datas. To quantify the strength of an transcription factor on a promoter, we will use measurements of GFP fluorescence, and compare to the strength of the constitutive promoter [J23101], as it was proposed by the iGEM competition.
The datas we are looking for must appear as a table of values, giving several 'transduction rates' with their corresponding 'transcription factor concentrations'.
first hypothesis
For this aim, we made several hypothesis, which we will verify as good as it is possible for us :
(1) We do not take into acount the 'traduction' phase, so we directly correlate the transcription of a gene with the concentration of its protein.
(2) We assume that, whatever is the gene behind the promoter, its expression depends only of the transcription factor of the promoter, and not, for instance, of the weight of this gene. That's why comparing promoter strength is relevent only if the genes behind have similar length.
(3) We consider that the activity of a promoter is well described as a Hill function of its transcription factor (TF).
Thus, we suppose that the protein concentration (Prot) follows this equation :
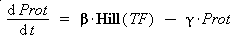
where gamma is a constant, due to degradation and of dilution of the protein, along time and cell divisions.
Therefore, if we consider a steady-state, for given concentration of the transcription factor, we will have :
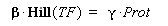
(4) Endly, knowing gamma will give us the kind of datas we are looking for. In a first approach, we assume that, as long as the barcteria are in their phase of exponential growth, the degradation is far smaller than the dilution, and can be omitted. But we will probably discuss that later.
(5) Unless we find further documents dealing with the relation between the intensity of fluorescence and the concentration of GFP, we will directly use the measure in fluorescence, that we will treat as a protein concentration, more or less arbitrary normalised.
how to control the concentration of the transcription factor ?
Now, we must use as a variable of reference an element that could be introduced in the bacteria, well-controlled, and from which will depends all the concentrations of our transcription factor. We propose a construction in which our transcription factor is put after the promoter Plac, which is under the repression of LacR. Since IPTG is a small diffusive molecule that binds to LacR and inhibits this way the repression of Plac, we can use it as an 'inducer'. To do so, we must place in the bacterium the gene lacR after a constitutive promoter (like J23101). According to previous hypothesis, this will provide at steady-state a 'constant concentration' of LacR (we note [LacR*], and it is supposed to be the TOTAL concentration of LacR, under every form) in the bacterium. If we consider the binding reaction this way (where LacR_IPTG denotes the complex)
LacR + IPTG ⇄ LacR_IPTG
with a dissociation constant K, we find at the steady-state
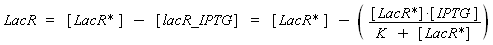
where [IPTG] denotes the concentration of IPTG we introduced in the medium, that will stay constant in all the bacteria along time, assuming that its degradation is near 0, and that the diffusion is quick. (We can already notice an other limitation in our protocol : this formula has got sense only for [IPTG] < (K + [LacR*]), that limits the range of [IPTG] we can use to determine the functions below. Notice that this limitation is conditionned by [LacR*], which depends of the constitutive promoter we put before lacR.)
According to the hypothesis (3), the activity of Plac would verify (keeping the same notations) :
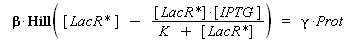
so, if k denotes the dissociation constant of (LacR + Plac ⇄ LacR_Plac),
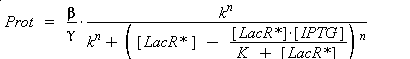
In the last equation, we will have 'access' (see hypothesis (5)
) to Prot and possibly to gamma, and we are looking for beta, k and n, thanks to our program (see getting Hill function with convenient datas). But we need to know ([LacR*] - [LacR*].[IPTG]/(K + [LacR*])), and we just have [IPTG]. However, we can reduce the last equation to
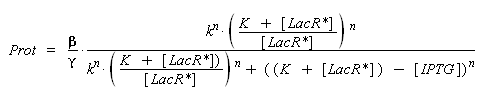
Thus, we have two possibilities :
- we can write a new algorithm that optimise an approaching solution of the new parameters, based on the same principal than 'findparam'.
- better but much longer and requiring much more precision, we can use the already noticed properties : [IPTG] < (K + [LacR*]). By having a look on the first equation of this section, we understand that beyond this limit, [LacR_IPTG] will no more evoluate. By observing the evolution of the influence of a growing (by steps as small as possible) concentration of [IPTG] introduced, we should be able to approximate the critic concentration when it no more changes, ~(K + [LacR*]). Less is the order n, better is the detection of this critic concentration, because of the greater derivative of the Hill function for small values of LacR. Therefore we should keep this estimation only if we find n ~ 1. Then, by considering k*((K + [LacR*])/[LacR*]) instead of k, we should easily determine all the parameters we need, only thanks to 'findparam'.
what are we looking for ?
The different functions we would like to determine are the followings. They are linked to a basic description of the 'experimental protocol' that will allow us to get the expected datas. We decide to let the original promoters in the bacteria, so that the strength that we are measuring is 'the strength for an additional promoter in the cell', keeping those who already exist.
According to the hypothesis (1) and (2), we assume this will directly give us [Protein] = ƒ1(IPTG), for a given Protein coded by a gene put behind the Plac promoter.
First Approach
As a first modeling, we have considered a set of five ordinary differential equations that are likely to represent the generic behaviour of the biological processes. The corresponding results and the associated code can be found there :
We will precise this description in the following part, entering into the details of the chemical reactions involved.
Moreover, once the oscillations have been obtained, it is interesting to focus on their robustness. To do such an analysis, we have used a rather intuitive algorithm which divides the parameter space into regular areas and compute a kind of 'score function' to test whether oscillations are observed. Then, for 2 parameters, a simple visualization is possible.
Therefore, the two aspects to describe a bit more are the following ones :
- There is a second score function, much simplier but quite efficient.
- We tried this one in association with a self made genetic algorithm, that allows us to find many convenient sets of parameters, in order to compare them.
More precise Bio-Mathematical Description
What kind of Mathematical Simulation ?
We decided to use mostly Ordinary Differential Equation approach, at least for the study of the Oscillations and of the FIFO. For the Synchronisation module, we will probably use Probabilistic Differential Equations, in order to introduce the differences between the cells.
Eventually our modellisation would just help us to find which ones of the sets of "biologicaly available parameters"/"logical circuit" are the bests, but it may help further simulations on similar protocols, too.
Bio-Chemical General Assumptions
We know that the following equations do not describe properly what really happens in the cells. For exemple, we know that the transcription factor flhD-flhC is actually an hexamere flhD4C2. But, as we will surely not get access to the dissociation constant of the hexamerisation, we just treat it as an abstract inducer protein flhDC, with an order in its Hill function probably between 3 and 6 (but perhaps completly different; the estimation of the error by the 'findparam' program will tell us if we are right to do so).
For the moment, at each part of our modelisation, we reduce the expression of a gene at its transcription. The translation process is not taken into acount.
To see more details about the modelisation and the values of the involved constants, see the bibliography.
Separated and detailed Parts of our Project
|
 "
"