A simple manner is to implement two class II promoter followed by fluorescent reporter genes like GFP in a bacteria strain that is able to produce the flagella.
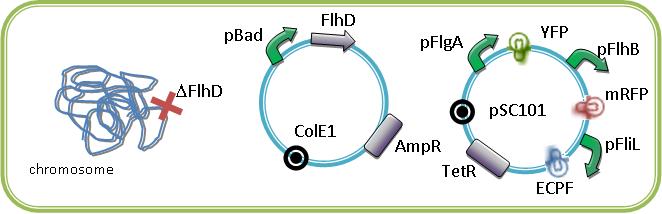
By this way, we are able to see in single cell the order of the activation of pFliL then pFlgA then pFlhB and then the inactivation in the same order as a FIFO will do.
These constructions are equivalent to the experiments realized by Uri Alon in the article "Using quantitative Blueprint to reprogram the Dynamics of the Flagella Gene Network" Kalir S, Alon U. Cell 2004.
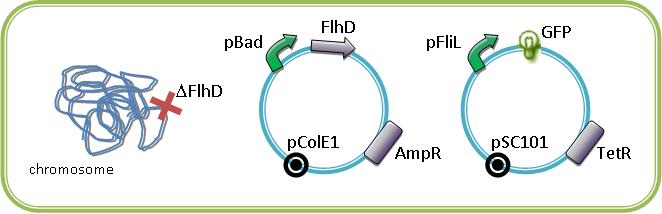
A better way to observe the FIFO is to study the system with an inducible regulator of class II gene in a bacteria strain deleted for this gene. We are extremely grateful for Alon U. that send us the inducible gene FlhDC, and FliA in pBad18 plasmid.
These plasmids with our own araC/pBad-EnvZ* will allowed us to study the system in mutated strains that we have in our lab (ΔFlhD, ΔFliA, ΔFlgM, ΔEnvZ). By cotransforming in mutated strains the inducible regulators of the class II promoters with one of this promoter associated to a fluorescent protein, we could characterize the influence of the master regulators of the flagella on their promoter. The fluorescence is normalized to the OD600.
For example below are some constructions to see and induce the FIFO:
We could see the FIFO in a more convenient manner with the assembly of two class II promoters associated to different fluorophores. Then we could observed two kinds of fluorescents in One Cell along time.
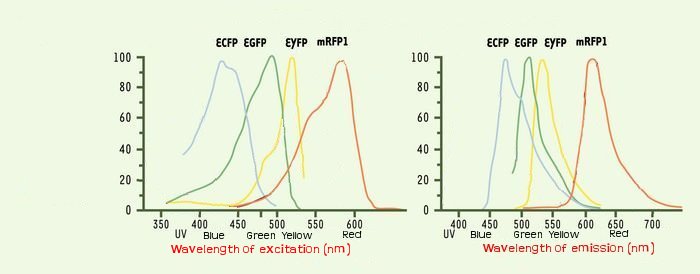
One problem that could occur is the interaction between the different fluorescents proteins due to the cross-over of their emission and excitation spectra and moreover in our final system that count three fluorophores.
We thank a lot Arnau Montagu, advisor of the iGEM Valencia Team. He advised us, after his team had troubleshooting with fluorescence, to take care about spurious FRET. This phenomenon occurs even with non covered fluorescent spectra when the concentration of two fluorophores is sufficient so they are enough closer to induce some FRET. But it could happen at lower concentration than 40 μM.
- "DsRed as a Potential FRET Partner with CFP and GFP" Michael G. Erickson, Daniel L. Moon, and David T. Yue Biophysical Journal 2003.
- "A Comparison of Donor-Acceptor Pairs for Genetically Encoded FRET Sensors: Application to the Epac cAMP
Sensor as an Example" Gerard N. M. van der Krogt, Janneke Ogink, Bas Ponsioen, Kees Jalink PLoS 2008.
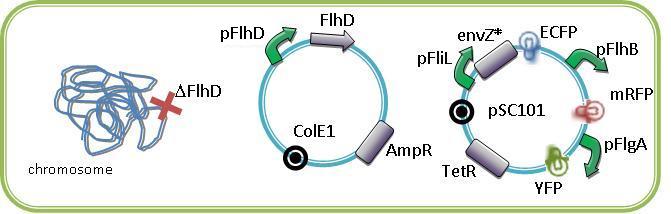
Few approaches have been thought in order to create the oscillating module. The pTet/TetR system that allows us to precisely control the inhibition of TetR on the Tet promoter. Another system use the natural inhibition of OmpR and of EnvZ via OmpR. We finally perform this system that we have developed since we know thanks to the modeling team that the oscillations are the only way to see the full development of the FIFO.
The modeling simulation showed us that to have some oscillations that are difficult to obtain in a simple feedforward loop without the delay introduce by the quorum sensing synchronization, and for us it is more interesting to have the FIFO behavior at the population level. We did not achieved this module but we started the construction of it and have already some intermediate parts that you will find below.
| FIFO
|
| Construction
| Description
| Validation Date
|
     
[http://partsregistry.org/Part:BBa_K136039 pFliL(RBS WT)-ECFP tripart (LVA+)]
| Natural Promoter of E. coli regulated by FlhDC and FliA. The natural rbs of the downstream gene is conserved for a more natural expression of the following gene. The fluorescent marker associated with pFliL is ECFP fused with a LVA tag.
| October 24
|
  
[http://partsregistry.org/Part:BBa_K136037 pFliL(+ RBS WT)-GFP]
| Natural Promoter of E. coli regulated by FlhDC and FliA. The natural rbs of the downstream gene is conserved for a more natural expression of the following gene. The fluorescent marker associated with pFliL is GFP.
| October 24
|
    
[http://partsregistry.org/Part:BBa_K136012 pFlgA(+ RBS WT) - YFP Tripart (LVA+)]
| Natural Promoter of E. coli regulated by FlhDC and FliA. The natural rbs of the downstream gene is conserved for a more natural expression of the following gene. The fluorescent marker associated with pFlgA is EYFP fused with a LVA tag.
| August 22
|
    
[http://partsregistry.org/Part:BBa_K136013 pFlgA(+ RBS WT) - YFP Tripart (LVA-)]
| Natural Promoter of E. coli regulated by FlhDC and FliA. The natural rbs of the downstream gene is conserved for a more natural expression of the following gene. The fluorescent marker associated with pFlgA is EYFP fused without a LVA tag.
| August 22
|
     
[http://partsregistry.org/Part:BBa_K136054 pFlgA(+ RBS WT)-YFP tripart (AAV+)]
| Natural Promoter of E. coli regulated by FlhDC and FliA. The natural rbs of the downstream gene is conserved for a more natural expression of the following gene. The fluorescent marker associated with pFlgA is EYFP fused with a AAV tag.
| October 24
|
  
[http://partsregistry.org/Part:BBa_K136053 pFlgA(+ RBS WT)-GFP]
| Natural Promoter of E. coli regulated by FlhDC and FliA. The natural rbs of the downstream gene is conserved for a more natural expression of the following gene. The fluorescent marker associated with pFlgA is GFP.
| October 24
|
     
[http://partsregistry.org/Part:BBa_K136055 pFlhB(+ RBS WT)-mRFP tripart (+LVA)]
| Natural Promoter of E. coli regulated by FlhDC and FliA. The natural rbs of the downstream gene is conserved for a more natural expression of the following gene. The fluorescent marker associated with pFlhB is mRFP with a LVA tag.
| October 24
|
  
[http://partsregistry.org/Part:BBa_K136065 pFlhB(+ RBS WT)-GFP]
| Natural Promoter of E. coli regulated by FlhDC and FliA. The natural rbs of the downstream gene is conserved for a more natural expression of the following gene. The fluorescent marker associated with pFlhB is GFP.
| October 26
|
| OSCILLATION
|
  
[http://partsregistry.org/Part:BBa_K136064 pFliL(+ RBS WT)-EnvZ*]
| pFliL is regulated by FlhDC and EnvZ regulates pFlhDC thanks to a negative feedback loop, origin of an oscillation behavior.
| October 26
|
      
[http://partsregistry.org/Part:BBa_K136069 pFliL(+ RBS WT)-EnvZ*-GFP generator]
| The same construction as before but with a GFP in order to follow the dynamic of expression of pFliL.
| October 29
|
  
[http://partsregistry.org/Part:BBa_K136051 pFlhB(+ RBS WT)-EnvZ*]
| pFlhB is regulated by FlhDC and EnvZ regulates pFlhDC thanks to a negative feedback loop, origin of an oscillation behavior.
| October 24
|
      
[http://partsregistry.org/Part:BBa_K136070 pFlhB(+ RBS WT)-EnvZ*-GFP generator]
| The same construction as before but with a GFP in order to follow the dynamic of expression of pFlhB.
| October 29
|
 
[http://partsregistry.org/Part:BBa_K136066 araC/pBad-EnvZ*]
| A construction made for characterizing the repression power of the mutated EnvZ on our system
| October 27
|
| SYNCHRONISATION
|
    
[http://partsregistry.org/Part:BBa_K136029 pLas - gfp Tripart]
| Construction for characterization of the pLas promoter
| August 1
|
    
[http://partsregistry.org/Part:BBa_K136027 pLas - ECFP]
| The same as before but with another fluorophore
| August 1
|
    
[http://partsregistry.org/Part:BBa_K136024 strongest promoter-rbs-lasR-dble ter]
| A construction for a strong constitutive expression of the Las Regulator in order to get maximal expression in presence of AHL
| August 20
|
    
[http://partsregistry.org/Part:BBa_K136023 Strong promoter-rbs-lasR-dble ter]
| A derived construction of the previous one for smaller constitutive expression of LasR in the Bacteria.
| August 20
|
 
[http://partsregistry.org/Part:BBa_K136044 Strongest RBS (1)- LasR activator (+LVA)]
| Intermediary construction. We uses different rbs for Las R in order to get various response dynamic to the presence of AHL
| August 13
|
   
[http://partsregistry.org/Part:BBa_K136048 rbs-lasR + dbl ter]
| Intermediary construction. We uses different rbs for Las R in order to get various response dynamic to the presence of AHL
| August 19
|
  
[http://partsregistry.org/Part:BBa_K136052 lasR activator + LVA - Double Terminator]
| Intermediary construction
| August 1
|
     
[http://partsregistry.org/Part:BBa_K136057 RBS lasI - GFP tripart]
| Construction that has to be associated with the FIFO. We will know when LasI is expressed and then when AHL is produced.
| August 13
|
     
[http://partsregistry.org/Part:BBa_K136056 RBS-lasI - ECFP]
| The same as the previous one but with another fluorophore.
| August 13
|
| NEW GENE AND PROMOTER BIOBRICKS
|

[http://partsregistry.org/Part:BBa_K136010 pFliA]
| Natural promoter of E. coli, extracted by PCR and then inserted in a biobrick vector. pFiA is regulated by FlhDC.
| October 04
|

[http://partsregistry.org/Part:BBa_K136009 pFliL (+RBS WT)]
| Natural promoter with the wild type rbs of its associated gene in E. coli, extracted by PCR and then inserted in a biobrick vector. pFiA is regulated by FlhDC and FliA.
| October 10
|

[http://partsregistry.org/Part:BBa_K136006 pFlgA (+RBS WT)]
| Natural promoter with the wild type rbs of its associated gene in E. coli, extracted by PCR and then inserted in a biobrick vector. pFiA is regulated by FlhDC and FliA..
| October 10
|

pFlgB (+RBS WT)
| Natural promoter with the wild type rbs of its associated gene in E. coli, extracted by PCR and then inserted in a biobrick vector. pFiA is regulated by FlhDC and FliA.
| October 10
|

[http://partsregistry.org/Part:BBa_K136008 pFlhB (+RBS WT)]
| Natural promoter with the wild type rbs of its associated gene in E. coli, extracted by PCR and then inserted in a biobrick vector. pFiA is regulated by FlhDC and FliA.
| October 10
|

[http://partsregistry.org/Part:BBa_K136046 EnvZ*]
| EnvZ is a membrane protein involved into the osmotic response and is interacting with OmpR that will bind the promoter of FlhDC ad will repress it. EnvZ* is a mutated form of the coding sequence, giving a phenotype where pFlhDC is completely repressed. EnvZ has been extracted by PVR for the correct mutant strain.
| September 04
|

FliA
| FliA is a sigma factor involved into the regulation of very various genes, the promoter used in the FIFO were selected to be higly responding to FliA. In order to have the FliA gene possibly used as biobrick, we mutated 2 sites (EcoRI and PstI) inside the FliA sequence by assemby PCR.
| September 04
|

FlhDC
| FlhDC is the main regulator of the flagella genetic system in E. coli. FlhDC regulates all the promoter of the FIFO.
| August 13
|
 "
"