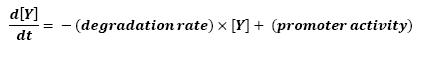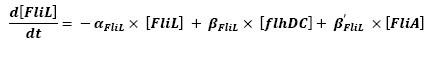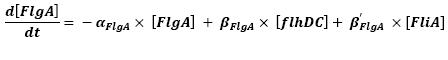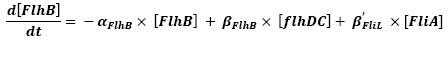An Oscillatory biological Model
Introduction
The goal here is to present the differential equations we used for our system modelization. At each step, we shall describe why we chose this precise model, its drawbacks and possible improvments, the parameters involved and enventually a biologically coherent value.
The key problem with a differential system consists in the fact that adding a new equation gives a more detailed idea of the overall process, but one looses precision by doing so since new parameters appear.
In that respect (in all cases but one) we chose not to model the mRNA steps, that is translation and transcription. We then assumed that we could act as if a protein would directly beget another one, without loosing too much precision.
Bibliography
We whall refer to those three articles :
[1] Shiraz Kalir, Uri Alon. Using quantitative blueprint to reprogram the dynamics of the flagella network. Cell, June 11, 2004, Vol.117, 713-720.
[2]Jordi Garcia-Ojalvo, Michael B. Elowitz, Steven H. Strogratz. Modeling a synthetic multicellular clock : repressilators coupled by quorum sensing. PNAS, July 27, 204, Vol. 101, no. 30.
[3]Nitzan Rosenfeld, Uri Alon. Response delays and the structure of transcription networks. JMB, 2003, 329, 645-654.
[4]Nitzan Rosenfeld, Michael B. Elowitz, Uri Alon. Negative autoregulation speeds the response times of transcription networks. JMB, 2003, 323, 785-793.
Equations
- FlhDC ---> FliL (1)
- FlhDC ---> FlgA (2)
- FlhDC ---> FlhB (3)
- FliA ---> FliL (4)
- FliA ---> FlgA (5)
- FliA ---> FlhB (6)
For all these equations, we found in [1] that in that precise case, the promoter activity the seven class 2 operons, among which FLiL, FlgA, FlhB, may be mathematically described in that way :
where [X] denotes the effective protein-level activity at time.
For each operon, Shiraz Kalir and Uri Alon came up with numerical values of β and β', available in [1].
Furthermore, the protein-level activity can be presented (for a more detailed presentation, see[4]) as
Thus :
Parameters summary
Graph screenshots
Roadmap
If you want to have a look at our roadmap : Roadmap
Bibliography
In order to choose a proper modeling approach for our system, we have decided to list all the chemical reactions we will take into account. Afterwards, we will find the needed parameters reading articles or devising the required experiments.
An overview of the work that has to be done can be found here : Bibliography
Estimation of parameters
Estimation of the parameters
First Approach
As a first modeling, we have considered a set of five ordinary differential equations that are likely to represent the generic behaviour of the biological processes. The corresponding results and the associated code can be found there :
We will precise this description in the following part, entering into the details of the chemical reactions involved.
Moreover, once the oscillations have been obtained, it is interesting to focus on their robustness. To do such an analysis, we have used a rather intuitive algorithm which divides the parameter space into regular areas and compute a kind of 'score function' to test whether oscillations are observed. Then, for 2 parameters, a simple visualization is possible.
Therefore, the two aspects to describe a bit more are the following ones :
- There is a second score function, much simplier but quite efficient.
- We tried this one in association with a self made genetic algorithm, that allows us to find many convenient sets of parameters, in order to compare them.
More precise Bio-Mathematical Description
What kind of Mathematical Simulation ?
We decided to use mostly Ordinary Differential Equation approach, at least for the study of the Oscillations and of the FIFO. For the Synchronisation module, we will probably use Probabilistic Differential Equations, in order to introduce the differences between the cells.
Eventually our modellisation would just help us to find which ones of the sets of "biologicaly available parameters"/"logical circuit" are the bests, but it may help further simulations on similar protocols, too.
Bio-Chemical General Assumptions
We know that the following equations do not describe properly what really happens in the cells. For exemple, we know that the transcription factor flhD-flhC is actually an hexamere flhD4C2. But, as we will surely not get access to the dissociation constant of the hexamerisation, we just treat it as an abstract inducer protein flhDC, with an order in its Hill function probably between 3 and 6 (but perhaps completly different; the estimation of the error by the 'findparam' program will tell us if we are right to do so).
For the moment, at each part of our modelisation, we reduce the expression of a gene at its transcription. The translation process is not taken into acount.
To see more details about the modelisation and the values of the involved constants, see the bibliography.
Separated and detailed Parts of our Project
|
 "
"