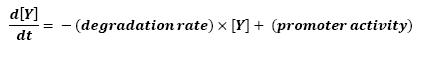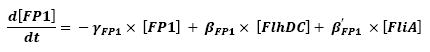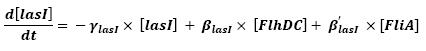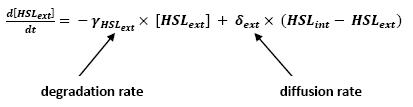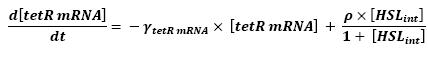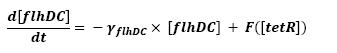Team:Paris/Modeling
From 2008.igem.org
(→An Oscillatory Biological Model) |
(→An Oscillatory Biological Model) |
||
| Line 50: | Line 50: | ||
===Equations=== | ===Equations=== | ||
| - | * flhDC → fliL → Fluorescent Protein 1 (FP1) ( | + | * flhDC → fliA (1) |
| - | * flhDC → flgA → Fluorescent Protein 2 (FP2) ( | + | * flhDC → fliL → Fluorescent Protein 1 (FP1) (2) |
| - | * flhDC → flhB → Fluorescent Protein 3 (FP3) ( | + | * flhDC → flgA → Fluorescent Protein 2 (FP2) (3) |
| - | * flhDC → flhB → lasI ( | + | * flhDC → flhB → Fluorescent Protein 3 (FP3) (4) |
| + | * flhDC → flhB → lasI (5) | ||
<br> | <br> | ||
| - | * fliA → fliL → Fluorescent Protein 1 (FP1) ( | + | * fliA → fliL → Fluorescent Protein 1 (FP1) (6) |
| - | * fliA → flgA → Fluorescent Protein 2 (FP2) ( | + | * fliA → flgA → Fluorescent Protein 2 (FP2) (7) |
| - | * fliA → flhB → Fluorescent Protein 3 (FP3) ( | + | * fliA → flhB → Fluorescent Protein 3 (FP3) (8) |
| - | * fliA → flhB → lasI ( | + | * fliA → flhB → lasI (9) |
<br> | <br> | ||
<br> | <br> | ||
| Line 73: | Line 74: | ||
<br> [[Image:equation1.jpg|center]] | <br> [[Image:equation1.jpg|center]] | ||
<br>Thus : | <br>Thus : | ||
| + | [[Image:fliA.jpg|center]] | ||
[[Image:FP1.jpg|center]] | [[Image:FP1.jpg|center]] | ||
[[Image:FP2.jpg|center]] | [[Image:FP2.jpg|center]] | ||
| Line 82: | Line 84: | ||
---- | ---- | ||
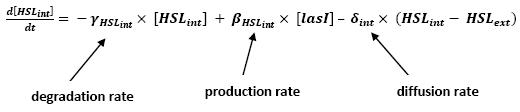
Even though we considered a single cell, we decided to model both HSL inside and outside the cell. In a first approach, we assumed that HSL could be modelized in the same fashion as AHL. The process was well detailed in [2]. | Even though we considered a single cell, we decided to model both HSL inside and outside the cell. In a first approach, we assumed that HSL could be modelized in the same fashion as AHL. The process was well detailed in [2]. | ||
| - | * lasI &rarr HSL<sub>ext</sub> ( | + | * lasI → HSL<sub>ext</sub> (10) |
| - | * lasI → HSL<sub>int</sub> ( | + | * lasI → HSL<sub>int</sub> (11) |
[[Image:HSLint.jpg|center]] | [[Image:HSLint.jpg|center]] | ||
| Line 90: | Line 92: | ||
---- | ---- | ||
| - | Here is the only case where we coul not afford to skip the mRNA step. Then, ( | + | Here is the only case where we coul not afford to skip the mRNA step. Then, (12) represents the binding and transcription steps ; (13) represents the translation step. |
| - | * HSL<sub>int</sub> → tetR mRNA ( | + | * HSL<sub>int</sub> → tetR mRNA (12) |
| - | * tetR mRNA → tet R ( | + | * tetR mRNA → tet R (13) |
hence : | hence : | ||
Revision as of 13:42, 6 August 2008
|
 "
"