Team:The University of Alberta/5 June 2008
From 2008.igem.org
(New page: ==To Do Today== *confirm Blue Ox Biobrick *plant plants *water plants in the growth chamber *colony PCR for all of the butanol biobricks in both J61003 and I0500 *read lots of papers! *fin...) |
|||
| Line 1: | Line 1: | ||
==To Do Today== | ==To Do Today== | ||
| - | *confirm Blue Ox Biobrick | + | *<strike>confirm Blue Ox Biobrick</strike> |
| - | *plant plants | + | *<strike>plant plants</strike> |
| - | *water plants in the growth chamber | + | *<strike>water plants in the growth chamber</strike> |
| - | *colony PCR for all of the butanol biobricks in both J61003 and I0500 | + | *<strike>colony PCR for all of the butanol biobricks in both J61003 and I0500 to check for postivie colonies</strike> |
| + | *set up O/N of positive colonies so we can do more protein stuff | ||
*read lots of papers! | *read lots of papers! | ||
*finish protein gel/begin Westerns(?) | *finish protein gel/begin Westerns(?) | ||
*<u>''meeting at 16:00''</u> | *<u>''meeting at 16:00''</u> | ||
| + | |||
| + | ==Our First BioBrick== | ||
| + | Ladies and Gentlemen, we have created our first BioBrick for the year. After getting a positive result with the EcoRI site experiment with the Blue Ox biobrick yesterday, we did the digestion and ran out the products again, to confirm that it did indeed work. Below is the result:<br> | ||
| + | [[Image:blue_ox_confirm.jpg]] | ||
| + | <br> | ||
| + | As you can see, there are two very distinct bands. This means that both the suffix and prefix are working properly. The Blue Ox is complete! | ||
| + | ''N.B'': You might notice that the band for Blue Ox is a fair bit larger than the expected 1111bp. This is because we originally cut out Blue Ox with XbaI and PstI, because the EcoRI site was faulty. We ligated it into J61003 which we had cut with the same enzymes. This gave Blue Ox a working EcoRI site in its prefix. However, for whatever reason, J61003 has a ~200bp spacer region in the prefix between the EcoRI and XbaI site. This is why Blue Ox appears to be larger than expected. Perhaps we should try to remove this region by ligating it into I0500 instead...nevertheless, we have our first BioBrick. | ||
| + | |||
| + | We may want to sequence the BioBrick again at a later point, just to be completely sure that it is what we think it is. | ||
| + | <br> | ||
| + | -[[User:Cwk|Cwk]] 20:27, 5 June 2008 (UTC) | ||
Revision as of 20:27, 5 June 2008
To Do Today
confirm Blue Ox Biobrickplant plantswater plants in the growth chambercolony PCR for all of the butanol biobricks in both J61003 and I0500 to check for postivie colonies- set up O/N of positive colonies so we can do more protein stuff
- read lots of papers!
- finish protein gel/begin Westerns(?)
- meeting at 16:00
Our First BioBrick
Ladies and Gentlemen, we have created our first BioBrick for the year. After getting a positive result with the EcoRI site experiment with the Blue Ox biobrick yesterday, we did the digestion and ran out the products again, to confirm that it did indeed work. Below is the result:
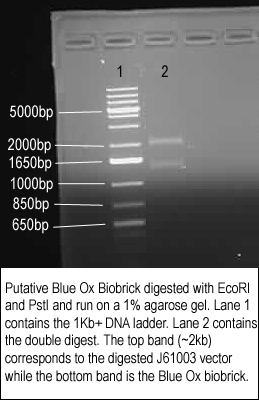
As you can see, there are two very distinct bands. This means that both the suffix and prefix are working properly. The Blue Ox is complete!
N.B: You might notice that the band for Blue Ox is a fair bit larger than the expected 1111bp. This is because we originally cut out Blue Ox with XbaI and PstI, because the EcoRI site was faulty. We ligated it into J61003 which we had cut with the same enzymes. This gave Blue Ox a working EcoRI site in its prefix. However, for whatever reason, J61003 has a ~200bp spacer region in the prefix between the EcoRI and XbaI site. This is why Blue Ox appears to be larger than expected. Perhaps we should try to remove this region by ligating it into I0500 instead...nevertheless, we have our first BioBrick.
We may want to sequence the BioBrick again at a later point, just to be completely sure that it is what we think it is.
-Cwk 20:27, 5 June 2008 (UTC)
 "
"