Team:The University of Alberta/Optimization Tutorial
From 2008.igem.org
(→Step 2) |
(→Step 6) |
||
| (7 intermediate revisions not shown) | |||
| Line 10: | Line 10: | ||
==Step 2== | ==Step 2== | ||
The second step is to choose which organism that you want to optimize your sequences for. We will be using E.Coli for the most part. The site is pretty specific, and you will have to chose which exact strain of E.coli that you want. The strain we are using is XL10 but its not on the list, so use K12 instead (I picked this one because its the strain on the list that I am most familiar with). Later in the year we may want to optimize sequences for use in Agrobacterium instead.<br> | The second step is to choose which organism that you want to optimize your sequences for. We will be using E.Coli for the most part. The site is pretty specific, and you will have to chose which exact strain of E.coli that you want. The strain we are using is XL10 but its not on the list, so use K12 instead (I picked this one because its the strain on the list that I am most familiar with). Later in the year we may want to optimize sequences for use in Agrobacterium instead.<br> | ||
| - | [[Image:opt2.jpg]]<br> | + | [[Image:opt2.jpg |600px]]<br> |
Once you have your organism of choice selected, press Continue. | Once you have your organism of choice selected, press Continue. | ||
==Step 3== | ==Step 3== | ||
| + | The next step is pretty simple. It will ask you to specify the genetic code you want to use. Pick eubacterial (this should be the default selection) and press continue. | ||
| + | Next it will ask you to specify which optimization method you want to use. Pick "One AA - one codon" (Should also be the default selection) to optimize each codon in the sequence. Press continue. | ||
| + | |||
==Step 4== | ==Step 4== | ||
| + | A new window should pop-up once the sequence has been optimized. The output should look similar to the one below:<br> | ||
| + | [[Image:opt3.jpg| 600px]]<br> | ||
| + | The page shows the sequence you input ("Query"), the optimized sequence ("Optimized"), the %GC and %AT content, and the number of different codons used ("ENc").<br> | ||
| + | Below, there should be a bunch of tools. The one we want is on the left. It will allow you to find restriction enzyme sites in the optimized sequence and then edit the sequence to remove them. Scroll through the list to find the first restriction enzyme you want to check and click on "Click here"<br> | ||
| + | [[Image:Opt4.jpg]] | ||
| + | |||
==Step 5== | ==Step 5== | ||
| + | What you should now be presented with is a new window, showing the optimized sequence and the location of the selected restricted enzyme sites (if present) indicated in red:<br> | ||
| + | [[Image:Opt5.jpg|600px]]<br> | ||
| + | |||
| + | Scroll back down to the restriction enzyme tool. The last enzyme you looked at should still be selected. Check the little box next to the "Click Here" button to tell the program to remove the selected restriction enzyme site from the sequence. <u>Make sure</u> to select "Modified" and not "Optimized", or else the program will apply the changes to the original optimized version of your sequence rather than the modified version with the restriction sites edited out. Do this each time you view or avoid a restriction site.<br> | ||
| + | [[Image:Opt6.jpg]] | ||
| + | |||
==Step 6== | ==Step 6== | ||
| + | When you have removed all the restriction enzyme sites (EcoRI, PstI, SpeI, and XbaI and NotI) from the optimized sequence, you can then paste it back into a Word document, attach the BioBrick prefix and suffix, and add the His-tag. Your BioBrick is not complete! | ||
Latest revision as of 20:57, 22 May 2008
UNDER CONSTRUCTION
This tutorial will show you how to use the Optimizer website to optimize our BioBricks so we get the best expression possible in E.Coli. The site is a little tricky to use, so hopefully this will make things a little clearer for you.
Contents |
Step 1
The first step is to copy your sequence to be optimized. For the purposes of this tutorial, I will assume that you have already trimmed your sequence (i.e you're working with only the ORF, you've removed any introns [if working with a eukaryotic gene; done for you if you're using a cDNA sequence] and you've cut out the UTRs). Paste your sequence into the box, make sure that DNA has been selected, and press Continue.
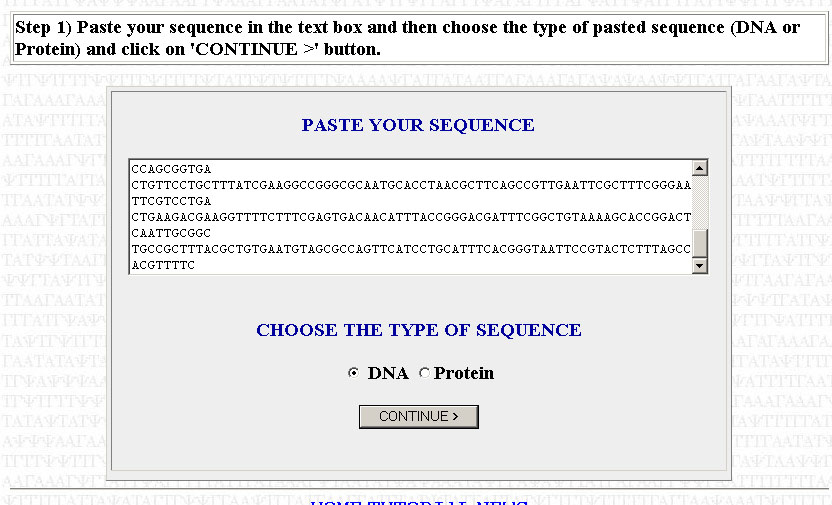
Make sure that your sequence is divisible by three or else it will not work. That is, each codon has to code for an amino acid; the sequence can contain no truncated codons. If you get an error telling you that your sequence is not divisible by three, then you need to go back to where you copied your sequence and check to see if you made a mistake in copying or if you added/subrtacted a nucleotide when trimming your sequence. Check to see if your trimmed sequence actually codes for the correct polypeptide by using the Translate tool posted in the May 16th notes. If the translated sequence is not what you expect, then you likely have a frameshift somewhere. You can use ClustalW (also posted in the May 16th notes to align your sequence with a reference sequence (from TAIR or NCBI) to find where the frameshift might be.
Step 2
The second step is to choose which organism that you want to optimize your sequences for. We will be using E.Coli for the most part. The site is pretty specific, and you will have to chose which exact strain of E.coli that you want. The strain we are using is XL10 but its not on the list, so use K12 instead (I picked this one because its the strain on the list that I am most familiar with). Later in the year we may want to optimize sequences for use in Agrobacterium instead.
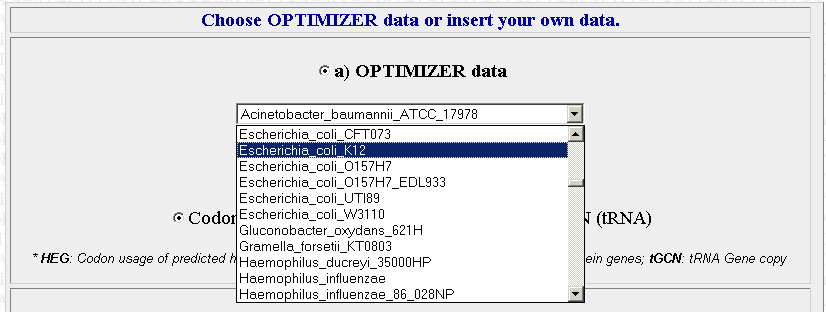
Once you have your organism of choice selected, press Continue.
Step 3
The next step is pretty simple. It will ask you to specify the genetic code you want to use. Pick eubacterial (this should be the default selection) and press continue. Next it will ask you to specify which optimization method you want to use. Pick "One AA - one codon" (Should also be the default selection) to optimize each codon in the sequence. Press continue.
Step 4
A new window should pop-up once the sequence has been optimized. The output should look similar to the one below:
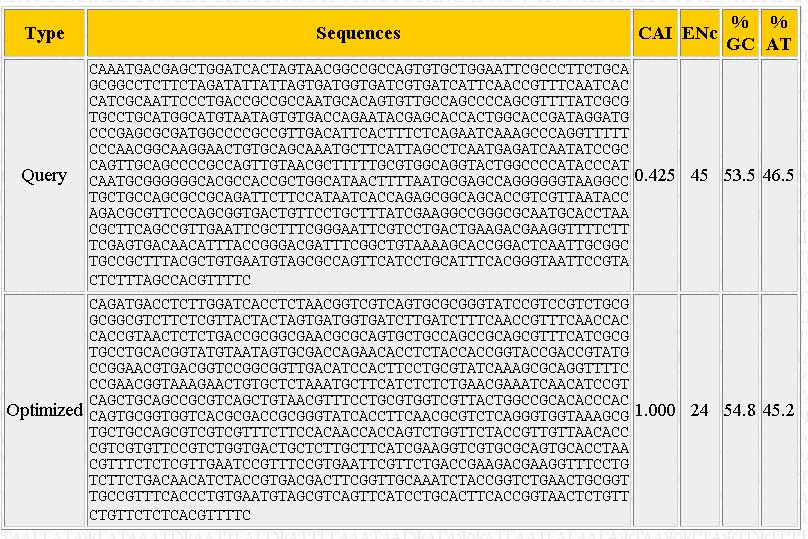
The page shows the sequence you input ("Query"), the optimized sequence ("Optimized"), the %GC and %AT content, and the number of different codons used ("ENc").
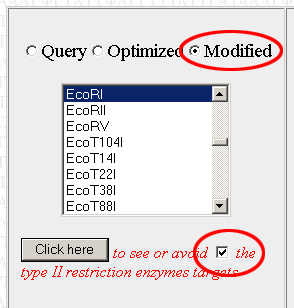
Below, there should be a bunch of tools. The one we want is on the left. It will allow you to find restriction enzyme sites in the optimized sequence and then edit the sequence to remove them. Scroll through the list to find the first restriction enzyme you want to check and click on "Click here"
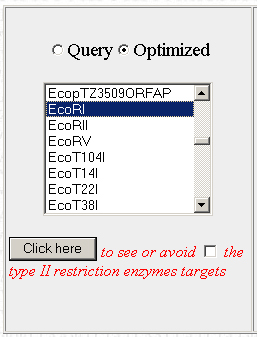
Step 5
What you should now be presented with is a new window, showing the optimized sequence and the location of the selected restricted enzyme sites (if present) indicated in red:

Scroll back down to the restriction enzyme tool. The last enzyme you looked at should still be selected. Check the little box next to the "Click Here" button to tell the program to remove the selected restriction enzyme site from the sequence. Make sure to select "Modified" and not "Optimized", or else the program will apply the changes to the original optimized version of your sequence rather than the modified version with the restriction sites edited out. Do this each time you view or avoid a restriction site.
Step 6
When you have removed all the restriction enzyme sites (EcoRI, PstI, SpeI, and XbaI and NotI) from the optimized sequence, you can then paste it back into a Word document, attach the BioBrick prefix and suffix, and add the His-tag. Your BioBrick is not complete!
 "
"