Imperial College/5 September 2008
From 2008.igem.org
5 September 2008WetlabCloning
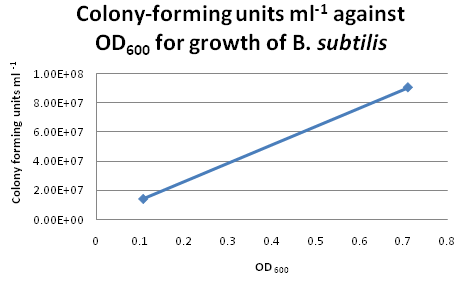
Colony CountingAfter re-calibrating our illuminator, it can now count colonies with a fair degree of accuracy. Manual addition and removal of points allows us to fine-tune the results, so we will use this method from now on. Trial Run: B. subtilis Growth CurveIn our second trial run to obtain growth and calibration curves for B. subtilis, we cultured our "B. subtilis" for two days and monitored the changes in OD600 continuously. By plotting OD600 against time/h, we obtained the following growth curve for "B. subtilis". Calibration CurveFor six of the data points from the above growth curve, we plated 0.1 ml of B. subtilis on LB agar plates and performed serial dilutions in order to calculate the colony-forming units (CFU per ml) for the corresponding OD600. However, we only managed to obtain two sets of valid data. The B. subtilis mixtures we plated for higher values of OD600 were too dilute, hence no bacterial growth was observed on these LB agar plates. Suggestions for Further ImprovementsFor the official experiment to obtain our calibration curve, we should take the following precautions:
Dry LabMotility
Microscopy2 microscopy sessions will be arranged for next week, on Wednesday and Friday.
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"