Team:Harvard/Dailybook/Week3/Chemical and Light
From 2008.igem.org
Goals for Week 3
[http://openwetware.org/images/d/dd/7-7monmtg_slides_%28compatible%29.ppt Summary Powerpoint by Amy]
The Cloning Strategy
Goals:
- Lac Plasmid: Hi/low promoter + RBS + LacI + terminator + pLac + RBS + GFP + terminator
- Tet Plasmid: Hi/low promoter + RBS + TetR + terminator + pTet + RBS + GFP + terminator
- cI Lambda Plasmid: Hi/low promoter + RBS + cI + terminator + pLambda + RBS + GFP + terminator
- Light Plasmid: P11 + ompC/ompF + RBS + GFP + terminator AND P11 + ompC/ompF + RBS
- CDF vector w/ different resistances: New Vector w/ CDF ori + Resistance Marker (and Death Gene)
Scoreboard: Keeping track of what we 'ave and don't 'ave
RBS + Repressor Coding Region
| Plasmid Number | Plasmid Description | Components Digested? | Components Purified? | Ligated? | Transformed? | Miniprepped? | Direct from Registry? |
| P56 | RBS + TetR (P40+P43) | Yes (7/1) | Yes (7/2) | Yes (7/2) | Yes, in E1 (7/2) | ||
| P57 | RBS + cI lambda (P40+P44) | Yes (7/1) | Yes (7/2) | Yes (7/2) | Yes, E1 (7/2) | ||
| RBS + LacI |
High/ Low Constitutive Promoters + RBS + Repressor Coding Region
| Plasmid Number | Plasmid Description | Components Digested? | Components Purified? | Ligated? | Transformed? | Miniprepped? | Direct from Registry? |
| P66 | High + RBS + TetR (P56 +P38) | ||||||
| P67 | Low + RBS + TetR (P56 + P39) | ||||||
| P68 | High + RBS + cI Lambda (P57 + P38) | ||||||
| P69 | Low + RBS + cI Lambda (P57 + P39) | ||||||
| High + RBS + LacI | |||||||
| Low + RBS + LacI |
High/ Low Constitutive Promoters + RBS + Repressor Coding Region + Terminator(s)
| Plasmid Number | Plasmid Description | Components Digested? | Components Purified? | Ligated? | Transformed? | Miniprepped? | Direct from Registry? |
| P70 | High + RBS + TetR + Term (P66 + ) | ||||||
| P71 | Low + RBS + TetR + Term (P67 + P39) | ||||||
| P72 | High + RBS + cI Lambda + Term (P68 + P38) | ||||||
| P73 | Low + RBS + cI Lambda + Term (P69 + P39) | ||||||
| High + RBS + LacI + Term | |||||||
| Low + RBS + LacI + Term |
RBS + GFP + Term
| Plasmid Number | Plasmid Description | Components Digested? | Components Purified? | Ligated? | Transformed? | Miniprepped? | Direct from Registry? |
| P45 | GFP only w/ RBS & terminator | N/A | N/A | N/A | N/A | Yes (7/1) | Yes |
p-lambda/pTet/pLac Promoter + RBS + GFP + Term
| Plasmid Number | Plasmid Description | Components Digested? | Components Purified? | Ligated? | Transformed? | Miniprepped? | Direct from Registry? |
| P15 | pTet + RBS + GFP + Term | N/A | N/A | N/A | N/A | Yes | Yes |
| P20 | pLac + RBS + GFP + Term | N/A | N/A | N/A | N/A | Yes | Yes |
| P74 | pLambda + RBS + GFP + Term (P18 + P45) | Yes | Yes |
pLambda/pTet/pLac Promoter + RBS + GFP + Term in Vector with p15a ori
| Plasmid Number | Plasmid Description | Components Digested? | Components Purified? | Ligated? | Transformed? | Miniprepped? | Direct from Registry? |
| P58 | pTet + RBS + GFP + Term in p15a ori (P1 + P15) | Yes (7/1) | Yes (7/2) | Yes (7/2) | Yes (7/2) | ||
| P59 | pLac + RBS + GFP + Term in p15a ori (P1 + P20) | Yes (7/1) | Yes (7/2) | Yes (7/2) | Yes (7/2) | ||
| P75 | pLambda + RBS + GFP + Term in p15a ori (P1 + P74) |
High/ Low Constitutive Promoters + RBS + Repressor Coding Region + Terminator(s) + pLambda/pTet/pLac Promoter + RBS + GFP + Term in Vector with p15a ori
| Plasmid Number | Plasmid Description | Components Digested? | Components Purified? | Ligated? | Transformed? | Miniprepped? | Direct from Registry? |
| High + RBS + TetR + Term + pTet + RBS + GFP + Term in p15a ori (P70 + P58) | |||||||
| Low + RBS + TetR + Term + pTet + RBS + GFP + Term in p15a ori (P71 + P58) | |||||||
| High + RBS + cI Lambda + Term + pLambda + RBS + GFP + Term in p15a ori (P72 + P75) | |||||||
| Low + RBS + cI Lambda + Term + pLambda + RBS + GFP + Term in p15a ori (P73 + P75) | |||||||
| High + RBS + LacI + Term + pLac + RBS + GFP + Term in p15a ori (+ P59) | |||||||
| Low + RBS + LacI + Term + pLac + RBS + GFP + Term in p15a ori (+ P59) |
Making a new CDF vector
CDF ori as an insert
| Plasmid Number | Plasmid Description | Components Digested? | Components Purified? | Ligated? | Transformed? | Miniprepped? | Direct from Registry? |
| P76 | modified P13 + P1 (CDF ori + p15a vector) | P1- 7/2, modified P13- 7/3 |
CDF ori + Resistance Cassettes as inserts
| Plasmid Number | Plasmid Description | Components Digested? | Components Purified? | Ligated? | Transformed? | Miniprepped? | Direct from Registry? |
| P79 | P76 + P48 (CDF ori on p15a vector + Cm Resistance) | P48- 7/3 | |||||
| P80 | P76 + P49 (CDF ori on p15a vector + Amp Resistance) | P49- 7/3 |
New Vector w/ CDF ori + Resistance Marker
| Plasmid Number | Plasmid Description | Components Digested? | Components Purified? | Ligated? | Transformed? | Miniprepped? | Direct from Registry? |
| P81 | P79 + P5 (CDF ori + Cm resistance on pSB3K3 vector) | ||||||
| P82 | P80 + P5 (CDF ori + Amp resistance on pSB3K3 vector) |
Transformation of Parts/ Plasmids into S1
Transformation of Lac/Tet + GFP into S1
7/3: 25 mL culture of S1 was grown for electroporation and transformation.
| Plasmid Name | uL DNA transformed | Plates grew? | Picked colonies? | Miniprepped? | Glycerol Stocks? |
| P58B | 8uL | many small colonies, no GFP | Yes | no growth | |
| P59B | 8uL | 5 medium-sized orangish-pinkish colonies, GFP + | Yes | yes | |
| P12 (from Christina) | 5 uL | lots of small, but pinkish colonies | Yes | yes | |
| P27 | 2uL | 9 small/medium orangish colonies, also many very small white-ish colonies | Yes | yes | |
| P28 | 2uL | many tiny colonies, no GFP | Yes | no growth | |
| P29 | 5uL | many tiny colonies, no Venus fluor | Yes | no growth | |
| P30 | 5uL | 2 small pinkish colonies, YFP+ | Yes | yes | |
| P31 | 8uL | lots of very small colonies, no Venus | Yes | ||
| P32 | 5uL | many (>50) medium orangish-pinkish colonies, no YFP | Yes | yes |
7/9: colonies picked and grown up in liquid culture
7/10: glycerol stocks made and samples miniprepped
Miniprep Results
| Sample ID | Date | Time | ng/ul | 260/280 | 260/230 |
| blank | 7/10/2008 | 3:51 PM | 0 | NaN | NaN |
| p38a 1 | 7/10/2008 | 3:53 PM | 121.91 | 2.01 | 2.02 |
| p38a 2 | 7/10/2008 | 3:54 PM | 152.32 | 1.98 | 1.82 |
| p39a 1 | 7/10/2008 | 3:56 PM | 167.45 | 1.93 | 1.91 |
| p39a 2 | 7/10/2008 | 3:57 PM | 173.56 | 1.92 | 1.9 |
| p12a | 7/10/2008 | 4:07 PM | 79.39 | 2.01 | 2.1 |
| p12b | 7/10/2008 | 4:08 PM | 39.97 | 1.88 | 0.93 |
| p27a | 7/10/2008 | 4:09 PM | 90.41 | 2 | 1.66 |
| p27b | 7/10/2008 | 4:10 PM | 84.75 | 1.98 | 2.16 |
| p30 1 | 7/10/2008 | 4:12 PM | 556.03 | 1.92 | 2.21 |
| p30 2 | 7/10/2008 | 4:12 PM | 673.6 | 1.9 | 2.17 |
| p32a | 7/10/2008 | 4:13 PM | 582.95 | 1.92 | 2.22 |
| p32b | 7/10/2008 | 4:15 PM | 249.7 | 1.97 | 2.26 |
| p59b 1 | 7/10/2008 | 4:16 PM | 74.7 | 2.08 | 1.52 |
| p59b 2 | 7/10/2008 | 4:17 PM | 100.66 | 1.99 | 2.14 |
Chemical 'n' Lite
Transformations/Minipreps of Parts from Registries
07/07/08
07/07: We transformed P42, P11, P51, P52, P77, P78, P17, Q01121, BBa_J06911, BBa_J06912 from both the 2007 and 2008 registries (P11 was available only from the 2008 registry). We also made a positive transformation control using pUC19 DNA that came with the TOP10 cells. We made three negative plate controls using mock transformed bacteria on Amp, Kan, and Cm plates. We used E1.
FAILED TRANSFORMATIONS
2007 -- P51 (Kan), Q01121 (Kan), P17 (Kan), P52 (Kan), P78 (Kan), P77 (Kan)
2008 -- P52 (Kan), P51 (Kan), P78 (Kan), P77 (Kan), Q01121 (Kan), P17 (Kan), P11 (Cm)
The Cm and Kan mock transformations (negative controls) also failed to grow.
Alarmingly, the E1 mock transformation (no DNA, neg control) grew on the LB Carb plates. All of the Carb plates had some colonies, so we only picked from those with significantly higher numbers than the neg control. 3 colonies each from BBa_J06911 & BBa_J06912 2007 (temp sensitive LacI system) were picked and grown.
Successful Transformations
2007: P84, P85 (BBa_J06911, BBa_J06912) -- miniprepped/glycerol stocks made
The Amp negative control colonies did not grow in liquid LB Amp. The Amp positive control did grow in liquid LB Amp (pUC19).
Nanodrop of Minipreps (07/09)
| Plasmid | Date | Time | ng/ul | 260/280 | 260/230 |
| p59 (S1) | 7/9/2008 | 4:23 PM | 84.44 | 1.9 | 2.13 |
| p30 (S1) | 7/9/2008 | 4:25 PM | 40.37 | 2.23 | 2.19 |
| p84a (E1) | 7/9/2008 | 4:26 PM | 291.42 | 1.96 | 2.23 |
| p84b (E1) | 7/9/2008 | 4:28 PM | 349.61 | 1.91 | 2.03 |
| p84c (E1) | 7/9/2008 | 4:29 PM | 237.09 | 1.92 | 1.98 |
| p85a (E1) | 7/9/2008 | 4:30 PM | 327.45 | 1.94 | 2.12 |
| p85b (E1) | 7/9/2008 | 4:32 PM | 331.99 | 1.95 | 2.1 |
| p18 (E) | 7/9/2008 | 4:33 PM | 135.09 | 1.97 | 1.81 |
| p45 (E) | 7/9/2008 | 4:34 PM | 296.22 | 1.94 | 1.99 |
| p49 (E) | 7/9/2008 | 4:35 PM | 614.33 | 1.91 | 2.16 |
Re-transformations 07/10/08
We retransformed P17, P26, P26, P42, P78, P88 from both the 2007 and 2008 registries. We made the following changes to the iGEM transformation protocol:
- Heat shock for 30s
- Incubate for 90min in 250 μL SOC medium
- Prewarm plates
In addition to the biobrick samples we also ran Amp and Carb positive controls (pUC19) and Carb and Kan negative controls (mock transformations).
07/11/08 Results
The Carb negative control had about 25 colonies and the Kan negative control was blank. Both of the Amp and Carb positive controls had lawns of bacteria. P26 2007 had 13 colonies and P26 2008 had 1 colony (both Amp plates we poured ourselves). P38+45 and P39+45 both had many colonies, but these were not fluorescent as expected (these were both on Carb plates but there were many more colonies than on the Carb negative control).
We made liquid cultures of all the successful transformations in LB Amp.
P88 failed to grow. However, the P17 and P42 plates had 1/2 colonies after another 24h. These plates were saved.
Part PCRs
07/07
More CDF was PCRed (w/ BioBrick adapting primers): 45μL PCR supermix, 1μL S1P13 (435ng/μL), 1μL CDF-F primer (20μM), 1μL CDF-R primer (20μM), 2μL water. Conditions: 5min @ 94°C → 35x[45s @ 94°C → 45s @ 58.7°C → 1m45s @72°C] → 5min @ 72°C → ∞ @ 4°C, heated lid
07/08
P11 and P17 from 2008 (left from punch), and P26, P38, P39, P42, P51, P52, P77, P78, Q01121 from 2007 plate were PCRed: 45μL PCR supermix, 1μL DNA, 1μL BBpfx primer (20μM), 1μL BBsfx (20μM), 2μL water. Conditions: 5min @ 94°C → 35x[45s @ 94°C → 45s @ 59°C → 1m45s @72°C] → 5min @ 72°C → ∞ @ 4°C, heated lid We're PCRing these b/c transformation for amplification has not been going smoothly.
07/10
Of the products from the previous days, Q01121 (P89), P51, P52, P17, P77, and CDF appeared to have bands. Since we wanted a higher concentration of DNA, we redid all the PCRs, with the aim of cutting out the PCRs that worked. Additionally, we lowered the annealing temperature with the hope that the PCRs that didn't work would work.
CDF
Reaction mix: 45μL PCR supermix, 1μL S1P13 (435ng/μL), 1μL CDF-F primer (20μM), 1μL CDF-R primer (20μM), 2μL water Conditions: 5min @ 94°C → 35x[45s @ 94°C → 45s @ 57°C → 1m38s @72°C] → 5min @ 72°C → ∞ @ 4°C, heated lid
Registry Parts
Reaction mix: 45μL PCR supermix, 1μL DNA, 1μL BBsfx primer (20μM), 1μL BBpfx primer (20μM), 2μL water
- P17, P26, P42, P51, P52, P77, P78, P89: 5min @ 94°C → 35x[45s @ 94°C → 45s @ 57°C → 1m38s @72°C] → 5min @ 72°C → ∞ @ 4°C, heated lid
- P11: 5min @ 94°C → 35x[45s @ 94°C → 45s @ 56°C → 4m47s @72°C] → 5min @ 72°C → ∞ @ 4°C, heated lid
- P88: 5min @ 94°C → 35x[45s @ 94°C → 45s @ 57°C → 2m38s @72°C] → 5min @ 72°C → ∞ @ 4°C, heated lid
- Rxns done in triplicate generally, with samples using DNA from 2008 punches and 2007 plates when available. P17 was only from 2007. P11, P88 were only from 2008.
Results (7/11)
Expected band sizes in bp indicated
After the gel extraction (standard kit protocol), the negative control (gel with no DNA) read 5.4 ng/μL and the positive control (a bright band from the ladder) was 8.3 ng/μL according to the NanoDrop.
RE digests
07/07/08
Repeated digests of P1, P18, P26, P38, P39, P45, P56, P57
| P1B | P18 | P26A | P38B | P39A | P45B | P56B | P57B | |
| DNA | 10 μL | 20 μL | 3 μL | 5 μL | 5 μL | 3 μL | 10 μL | 10 μL |
| 10X NEB Buffer | buffer 3, 2.5 μL | buffer 2, 2.5 μL | buffer 3, 2.5 μL | buffer 2, 2.5 μL | buffer 2, 2.5 μL | buffer 3, 2.5 μL | buffer 3, 2.5 μL | buffer 3, 2.5 μL |
| 25X BSA | 1 μL | 1 μL | 1 μL | 1 μL | 1 μL | 1 μL | 1 μL | 1 μL |
| REs (1 μL each) | XP | SP | XP | SP | SP | XP | XP | XP |
| Water | 9.5 μL | 0 μL | 16.5 μL | 14.5 μL | 14.5 μL | 16.5 μL | 9.5 μL | 9.5 μL |
07/08: Digested pCDF with XP:
- 12.5 μL water
- 1 μL 25X BSA
- 2.5 μL 10X NEB Buffer 3
- 7 μL DNA
- 1 μL of each RE (XbaI and PstI)
07/08: All of these digested plasmids were run on a 1% low melt gel.
MXH's gel extraction protocol:
- Cut out the bands
- Freeze at –20C for at least 20'
- Spin 13000 RPM 10’
- Use supernatant for ligation
- Then do gel extraction on rest of gel (QIAGEN protocol) and combine with above
Here is a ligation protocol (used by TA and MXH):
- 2 μL 5X Dilution Buffer
- 2 μL vector
- 6 μL insert
- 10 μL 2X Rapid Ligation Buffer
- 1 μL ligase
- vortex briefly
- incubate at RT for 10'
- use 5 μL to transform 50 μL cells
07/08/08
We attempted to digest the following plasmids (per the Master Plan sent out to the iGEM google group):
- A (BBa_Q01121) with ES
- P11 with ES
- P17 with ES
- P23 with EX
- P24 with EX
- P26 with XP
- P38A with ES
- P38B with XP
- P39A with ES
- P39B with XP
- P42 with XP
- P56 with EX
- P58 with EX
- P59 with EX
- P63 with ES
- P77 with ES
- P78 with ES
These digests were run on gels and the bands of the correct size (when present) were extracted. If indicated, we digested PCR products of the plasmid as well (we did not purify PCR product first).
Gels (expected sizes of parts indicated)
1.5% gel (all of PCR products)
- 100 bp ladder
- A (BBa_Q01121) with ES: 1372 (NO BAND)
- P17 with ES: 902 (NO BAND)
- P26 with XP: 961 (NO BAND)
- P42 with XP: 1128 (NO BAND)
- P77 with ES: 1307 (NO BAND)
- P78 with ES: 1310 (NO BAND)
- CDF with XP: ~900 (NO BAND)
07/10/08 and 07/11/08
Digested P58 and 59 with EX, P45 with XP, P53 and P54 with ES, and P38 and P39 with SP. We plan to ligate:
- P53/54 with P58/59. This will give us RBS + LacI/TetR + terminator + pLac/pTet + RBS + GFP + terminator.
- P38/39 with P45. This will give us Hi/low constitutive promoter + RBS + GFP + terminator. This is to test whether we actually have P38 and P39.
These digests did not show bands of the right size on a gel and some of the lanes were blank. We re-did these digests in two ways: (1) using the same conditions as above (1 μL RE) and (2) using the NEB minimum required amount of enzyme (this was << 1 μL so we touched a pipette tip into to enzyme tube and swirled it around in the reaction tube). We saw bands of the correct size for P38 and P39 using the old and new methods, and bands of the correct size for P45 and P54 using the old method (1 μL enzyme). All of the other lanes (using both the old and new methods) did not have bands of the correct size.
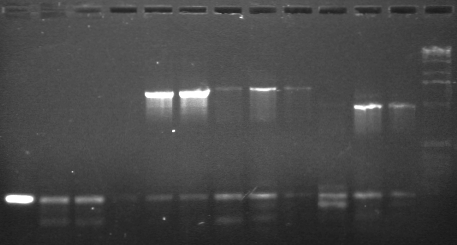
Lane 1: P38 SP (3037 bp) -- new
Lane 2: P38 SP (3037 bp) -- old
Lane 3: P39 SP (3037 bp) -- new
Lane 4: P39 SP (3037 bp) -- old
Lane 5: P45 XP (876 bp) -- new
Lane 6: P45 XP (876 bp) -- old
Lane 7: P53 ES (840 bp) -- new
Lane 8: P53 ES (840 bp) -- old
Lane 9: P54 ES (1308 bp) -- new
Lane 10: P54 ES (1308 bp) -- old
Lane 11: blank
Lane 12: blank
Lane 13: 1 kb ladder
7/11/08 Digests of Parts to Put into p15a Vector
| ' | P1 (vector) | P1 (vector) | P74 (insert) | P84 (insert) | P85 (insert) |
| DNA | 15 uL | 15 uL | 5 uL | 5 uL | 5 uL |
| 10X Buffer (Volume & #) | 2.5 uL of 3 | 2.5 uL of 3 | 2.5 uL of 3 | 2.5 uL of 3 | 2.5 uL of 3 |
| 100X BSA | 0.25 uL | 0.25 uL | 0.25 uL | 0.25 uL | 0.25 uL |
| Restriction Enzyme 1 | 1 uL XbaI | 1 uL XbaI | 1 uL XbaI | 1 uL XbaI | 1 uL XbaI |
| Restriction Enzyme 2 | 1 uL PstI | 1 uL PstI | 1 uL PstI | 1 uL PstI | 1 uL PstI |
| Water | 5.25 uL | 5.25 uL | 15.25 uL | 15.25 uL | 15.25 uL |
| Total Volume | 25 uL | 25 uL | 25 uL | 25 uL | 25 uL |
7/12: Digests of 7/11 PCR products, P45
Using the PCR products from 7/11 (gel purified and extracted w/ kit), the following digests were performed:
Reaction mixes: 10 μL DNA, 1 μL BSA, 1 μL enzyme mix, 2.5 μL buffer, 10.5 μL water (total volume: 25 μL) Reactions at 37°C for 2 hours. ES mix used EcoRI buffer, XP mix used buffer 3
P17 with ES P17 with XP P51 with XP P52 with XP P77 with ES P77 with XP P89 with ES P89 with XP CDF with XP
The above was purified using the Qiagen PCR purification kit (removes DNA <40bp, so the end fragments from the PCR should be removed).
Using the E1 P45B 7/11 miniprep DNA, a P45 EX digest was also performed. Reaction mix: 4 μL DNA, 1 μL BSA, 1 μL enzyme mix, 2.5 μL buffer 2, 16.5 μL water (total volume: 25 μL). Reactions at 37°C for 2 hours. Although this had the PCR purification procedure done, it still needs to be gel purified.
Ligations
mtrA and p45
7/7/08 Ligated mtrA and p45 using standard protocol from ligation kit. In step 2 5uL of Vector DNA was added to make a 20uL dephosphorylation mix. In step 6, 4uL of the dephosphorylated Vector DNA and 4uL insert DNA were added.
07/09/08: P59+63, P58+63, P18+45
We ligated P59 (vector) and P63 (insert); P58 (vector) and P63 (insert); P18 (vector) and P45 (insert).
- 2 μL 5X Dilution Buffer
- 2 μL vector
- 6 μL insert
- 10 μL 2X Rapid Ligation Buffer
- 1 μL ligase
- vortex briefly
- incubate at RT for 10'
- use 5 μL to transform 50 μL competent cells
We transformed the ligated plasmids (P59+63 and P58+63 on Kan, P18+45 on Amp). We also an Amp positive control of pUC19 and two mock transformations (negative controls).
Only the ligation of P18 to P45 (P74) was successful (the plate had colonies that were fluorescent). We made a glycerol stock and a LB amp culture.
Miniprep from Ligation and Glycerol Stocks (7/11/08)
| Sample ID | Date | Time | ng/ul | 260/280 | 260/230 |
| P74A | 7/11/2008 | 12:57 PM | 358.06 | 1.97 | 2.33 |
| P74B | 7/11/2008 | 12:58 PM | 578.78 | 1.91 | 2.27 |
| P74C | 7/11/2008 | 12:59 PM | 365.28 | 1.98 | 2.27 |
| P55B 2007 A | 7/11/2008 | 1:00 PM | 382.22 | 1.96 | 2.36 |
| P55B 2007 B | 7/11/2008 | 1:01 PM | 339.99 | 1.97 | 2.37 |
| P45B 2007 A | 7/11/2008 | 1:02 PM | 185.19 | 2.1 | 2.44 |
| P45B 2007 B | 7/11/2008 | 1:03 PM | 302.11 | 2.01 | 2.37 |
07/10/08
We used the standard ligation protocol to ligate P38 and P39 with P45. This will test the P38 and P39 samples that we have. P38 and P39 are the hi and low constitutive promoters; P45 is RBS + GFP + terminator. So if the colonies fluoresce, then our minipreps of P38 and P39 do contain P38 and P39.
7/12/08
The following were ligated, with 5μL used to transform E1:
Mutant Strand Syntheis Reaction
7/8/08 Used Stratagene QuikChange Site-Directed Mutatgenesis Kit [http://openwetware.org/wiki/IGEM:Harvard/2008/Lab_Notebooks/GenProtocols#Protocol_for_Mutant_Strand_Synthesis_Reaction (protocol)] to create a point mutation in the pstI site in mtrA. Used 5ug of primer.
Gradient PCR
7/8/08 Used gradient PCR to PCR mtrB. Cycled 30-40°C x 10 and then 52-62°C x 30.
07/10/08
- pPL-PCB from UT Austin successfully transformed into Shewanella (p15a ori)
- pCph1-envZ-pprotet did not transform into Shewie (ColE1 ori)
- The ligated p45-mtrA plasmid did not successfully mutate and/or transform into the ultracompetetent cells. This implies that we should focus our efforts on mtrB as it does not require a mutation.
Cross Transformations in S1
| Plate | Marker | Description | Picked Colonies? |
| S1 P34 | Amp | Lawn, restreaked. Many orange colonies along streak. Can\'t detect fluor. | Yes |
| S1 P59b cells + P12 vector | Amp | Lawn, restreaked. Fluorescent. ? | Yes |
| S1 P13 cells + P59b vector (1) | Kan | Many small colonies-- some fluoresce while others don\'t. Pinkish centers. | Yes (both F and no F) |
| S1 P13 cells + P59b vector (2) | Kan | More colonies than (1), but also some fluoresce while others don\'t. Pinkish centers. | Yes (both F and no F) |
| S1 P27 cells + P59B vector (1) | Kan | Many small colonies - fluorescent. | Yes |
| S1 P27 cells + P59B vector (2) | Kan | Almost lawn of tiny colonies - fluorescent. | Yes |
| S1 P59b Cells + P13 vector | Sm | Medium # of medium sized colonies, pink centers, no fluorescence. | Yes |
 "
"