Team:Harvard/Dailybook/Week6/Chemical and Light
From 2008.igem.org
Sequencing Parts
Making Thermoinducible cI System
Minipreps from Transformations of P75 + P63 7/28
| Sample ID | ng/uL | A260 | 260/280 | 260/230 | Constant |
| E1 P3+mtrB notBB A | 60.36 | 1.207 | 1.95 | 1.99 | 50 |
| E1 P3+mtrB notBB B | 53.82 | 1.076 | 1.98 | 1.92 | 50 |
| E2 P1+mtrB A | 98.14 | 1.963 | 1.94 | 1.83 | 50 |
| E2 P1+mtrB B | 119.82 | 2.396 | 1.91 | 1.96 | 50 |
| E2 P3+mtrB NotBB A | 113.02 | 2.26 | 1.97 | 1.88 | 50 |
| E2 P3+mtrB NotBB B | 122.34 | 2.447 | 1.98 | 2.07 | 50 |
| PGW7 1A | 69.69 | 1.394 | 1.91 | 1.91 | 50 |
| PGW7 1B | 81.88 | 1.638 | 1.86 | 1.86 | 50 |
| PGW7 2A | 88.69 | 1.774 | 1.92 | 1.83 | 50 |
| PGW7 2B | 56.57 | 1.131 | 1.82 | 1.85 | 50 |
| P104A (S1 P75A+P63) | 66 | 1.32 | 1.74 | 0.81 | 50 |
| P104B (S1 P75B+P63) | 52.58 | 1.052 | 1.71 | 0.93 | 50 |
Digestion of Term + pLambda GFP System and cI857 7/28
| ' | P104a | cI857 RBS | cI857 BIG |
| DNA | 15 uL | 13 uL | 20 uL |
| 10x Buffer | 2.5 uL | 2.5 uL | 2.5 uL |
| 100x BSA | 0.25 uL | 0.25 uL | 0.25 uL |
| Restriction Enzyme 1 | 1 uL EcoRI | 1 uL EcoRI | 1 uL EcoRI |
| Restriction Enzyme 2 | 1 uL XbaI | 1 uL SpeI | 1 uL SpeI |
| Water | 5uL | 7uL | 0uL |
| Total | 25uL | 25uL | 25uL |
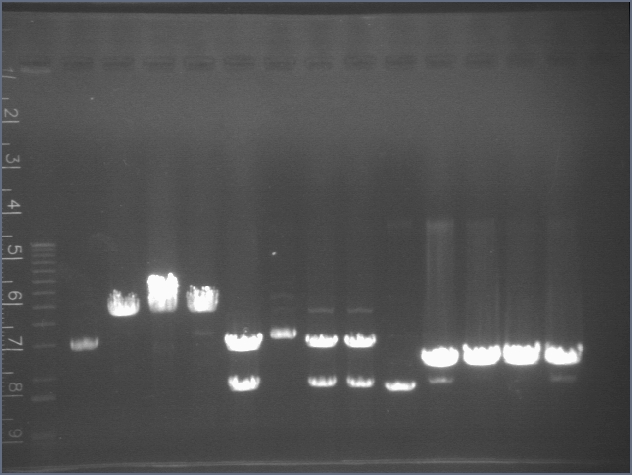
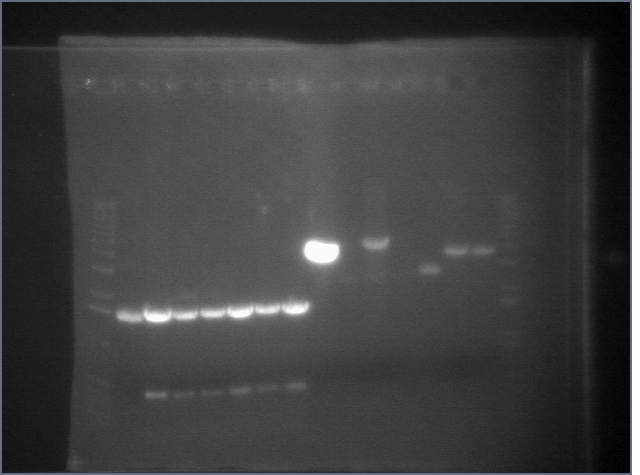
Gel of Digestion 7/29
Gel here-- P104 was cut correctly but I made a mistake and thought it hadn't been cut properly, so discarded gel and repeated digest.
Digestion of P104a with new XbaI 7/29
| ' | P104a |
| DNA | 20 uL |
| 100x BSA | 0.25 uL |
| 10x Buffer | 2.5 uL |
| EcoRI | 0.5 uL |
| XbaI | 1 uL |
| Water | 0.75 uL |
| Volume | 25 uL |
Gel for Digestion 7/30
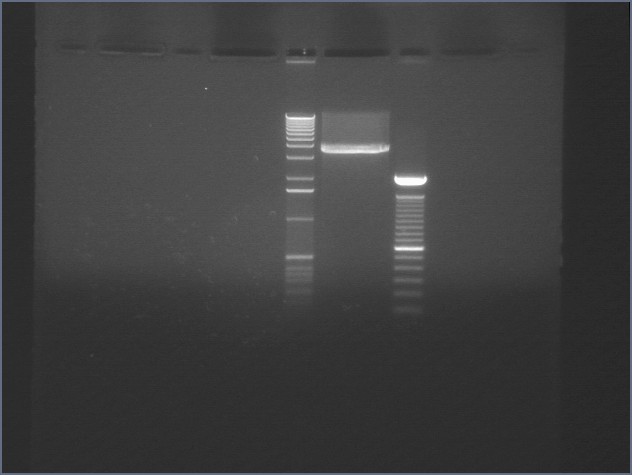
| 1% Agarose, visualized using EtBr/UV | |
|---|---|---|
| Lane | Sample | |
| 1 | 1 kB Ladder | |
| 2 | P104a (3675bp) | |
| 3 | 100bp Ladder | |
P104 was extracted and gel purified. It was dephosphorylated with the rAPid Alkaline Phosphatase.
Ligation of P104 and cI 7/30
| ' | P104a + RBS | P104a + BIG | P104a + RBS | P104a + BIG |
| Insert | 6 | 6 | 5 | 5 |
| Vector | 2 | 2 | 1 | 1 |
| Dilution Buffer | 2 | 2 | 2 | 2 |
| Water | 0 | 0 | 2 | 2 |
| Total | 10 | 10 | 10 | 10 |
| Then add: | ||||
| Ligation Buffer | 10 | 10 | 10 | 10 |
| Ligase | 1 | 1 | 1 | 1 |
| Total | 21 uL | 21 uL | 21 uL | 21 uL |
Transformed E1 with 5 uL of each ligation reaction.
Plate Results 7/31
| Plate | Marker | # Colonies | Description |
| P104a + RBS (6:2) | Kan | 0 | |
| P104a + BIG (6:2) | Kan | 0 | |
| P104a+ RBS (5:1) | Kan | 0 | |
| P104a + BIG (5:1) | Kan | 0 | |
| (+) Ctrl (puc19) | Amp | 25-50 | Not very evenly distributed. |
| (-) Ctrl (EB buffer) | Kan | 0 |
Re-Ligation of P104a + cI 7/31
Used the same purified fragments as before and ligated again in TOP10 cells.
| ' | P104a + RBS | P104a + BIG |
| Insert | 6 | 6 |
| Vector | 2 | 2 |
| Dilution Buffer | 2 | 2 |
| Water | 0 | 0 |
| Total | 10 | 10 |
| Then add: | ||
| Ligation Buffer | 10 | 10 |
| Ligase | 1 | 1 |
| Total | 21 | 21 |
Plate Results 8/1
| Plate | Marker | # Colonies |
| P104a + RBS | Kan | 0 |
| P104a + BIG | Kan | 0 |
| (+) Ctrl (puc19) | Amp | 200 |
| (-) Ctrl (EB buffer) | Kan | 0 |
Re-doing Cloning with P75, P63, cI, and P104a 7/31
PCR of PGW7 to get cI 7/31
| ' | cI857 BIG | cI857 RBS |
| Fwd Primer | 2 uL | 2 uL |
| Reverse Primer | 2 uL | 2 uL |
| PCR Supermix | 90 uL | 90 uL |
| Template | 2 uL | 2 uL |
| Water | 4 uL | 4 uL |
| Total | 100 uL | 100 uL |
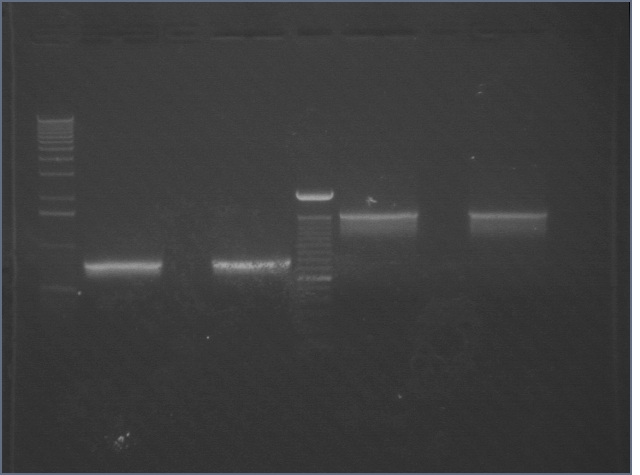
| 1% Agarose, visualized using EtBr/UV | |
|---|---|---|
| Lane | Sample | |
| 1 | 1 kB Ladder | |
| 2 | RBS 1 | |
| 3 | RBS 2 | |
| 4 | 100bp Ladder | |
| 5 | BIG 1 | |
| 6 | BIG 2 | |
Re-digestion of P75, P63, cI, and P104a 7/31
| ' | E1 P63b 07 | E1 P75a | E1 P75b | P104a |
| DNA | 30 uL | 10 uL | 10 uL | 9 uL |
| 100x BSA | 0.5 uL | 0.25 uL | 0.25 uL | 0.25 uL |
| 10x Buffer 2 | 5 uL | 2.5 uL | 2.5 uL | 2.5 uL |
| EcoRI | 1 uL | 2 uL | 3 uL | 4 uL |
| RE 2 | 1 uL SpeI | 1 uL XbaI | 1 uL XbaI | 1 uL XbaI |
| Water | 12.5 uL | 10.25 uL | 10.25 uL | 10.25 uL |
| Volume | 50 uL | 25 uL | 25 uL | 25 uL |
Gels from Digestions 8/1
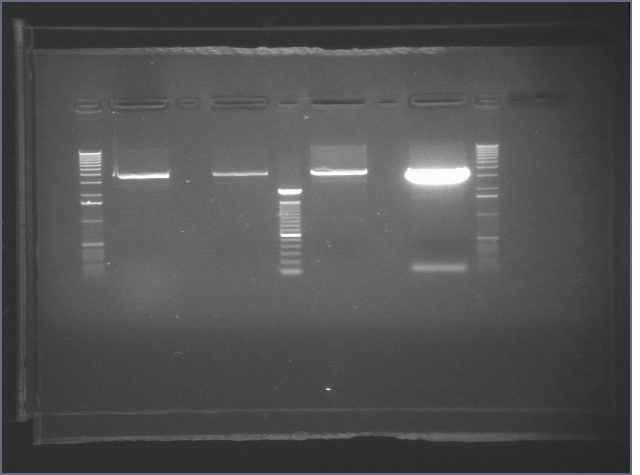
| 1% Agarose, visualized using EtBr/UV | |
|---|---|---|
| Lane | Sample | |
| 1 | 1 kB Ladder | |
| 2 | E1 P75a | |
| 3 | E1 P75b | |
| 4 | 100bp Ladder | |
| 5 | E1 P104 | |
| 6 | E1 P63b | |
Gels of Ligation Reactions 7/31
Ligation of P75 and P63 8/1
| ' | E1 P75a + P63b | E1 P75b + P63b | E1 P75a + P63b | E1 P75b + P63b | E1 P75a + P63b | E1 P75b + P63b |
| Insert | 6 uL | 6 uL | 5 uL | 5 uL | 1 uL | 1 uL |
| Vector | 2 uL | 2 uL | 1 uL | 1 uL | 1 uL | 1 uL |
| Dilution Buffer | 2 uL | 2 uL | 2 uL | 2 uL | 2 uL | 2 uL |
| Water | 0 uL | 0 uL | 2 uL | 2 uL | 6 uL | 6 uL |
| Total | 10 uL | 10 uL | 10 uL | 10 uL | 10 uL | 10 uL |
| Then add: | ||||||
| Ligation Buffer | 10 uL | 10 uL | 10 uL | 10 uL | 10 uL | 10 uL |
| Ligase | 1 uL | 1 uL | 1 uL | 1 uL | 1 uL | 1 uL |
| Total | 21 uL | 21 uL | 21 uL | 21 uL | 21 uL | 21 uL |
Transformed in E1.
Plates from Transformations 8/2
| Plate | Marker | # Colonies | Description |
| E1 P75a + P63b | Kan | ||
| E1 P75b + P63b | Kan | ||
| E1 P75a + P63b | Kan | ||
| E1 P75b + P63b | Kan | ||
| E1 P75a + P63b | Kan | ||
| E1 P75b + P63b | Kan | ||
| puc19 + ctrl | Amp | ||
| (-) ctrl | Kan |
TOPO TA cloning of cI
TOPO TA cloning with gel extracted cI 8/1
| ' | Volume |
| PCR Product | 0.5 - 4 uL |
| Salt Solution | 1 uL |
| TOPO Vector | 1 uL |
| Water | add to 6 uL |
Transformed in E1.
Plate Results 8/2
Includes results for ligation of P75a and P75b with P63 (terminator). RBS indicates primer set RBS, and BIG indicated primer set BIG. These TOPO cloning reactions were done with gel purified cI857. Volumes of DNA in parentheses indicate the amount of DNA added to the cloning reaction (0.5, 2, or 4 uL).
| Plate | Marker | # Colonies | Description |
| E1 P75a + P63b (1:1) | Kan | 0 | |
| E1 P75b + P63b (1:1) | Kan | 0 | |
| E1 P75a + P63b (6:2) | Kan | 1 | No Fluor, 3mm |
| E1 P75b + P63b (6:2) | Kan | 1 | Fluorescent, 1mm. |
| E1 P75a + P63b (7:1) | Kan | 0 | |
| E1 P75b + P63b (7:1) | Kan | 0 | |
| Dephos P75b | Kan | 0 | |
| RBS (0.5 uL DNA) | Amp | 24 big, 5 small | White, bi: 2mm. Smaller ones flanking larger colonies: <1 mm. |
| RBS (2 uL DNA) | Amp | ~40 big, 10-20 small | White, big colonies: 2mm. Smaller ones flanking them: <1mm. |
| RBS (4 uL DNA) | Amp | ~50 big, 10-20 small. | White, big colonies: 2mm. Smaller ones flanking them: <1mm. |
| BIG (0.5 uL DNA) | Amp | ~15 big | White, big colonies: 2mm. |
| BIG (2 uL DNA) | Amp | ~25 big | White, big colonies: 2mm. |
| BIG (4 uL DNA) | Amp | ~40 | White, big colonies: 2mm. |
| mtrB BB (0.5 uL DNA) | Amp | ~40 | White colonies, 2mm. |
| mtrB BB (2 uL DNA) | Amp | ~40-50 big, ~50 small | White, big colonies: 2mm. Smaller ones flanking them: <1mm. |
| mtrB BB (4 uL DNA) | Amp | ~20 big, ~20 small | White, big colonies: 2mm. Smaller ones in clusters: <1 mm. |
| puc19 + ctrl | Amp | 100 white, big colonies, 50+ small. | White, big colonies: 2mm. Smaller ones in clusters: <1 mm. |
| (-) ctrl | Kan | 0 | |
| just TOPO vector | Amp | ~25 big | White, big colonies: 1-2mm. |
| (-) ctrl | Amp | 0 |
PCR of PGW7 to get more cI 8/1
| ' | cI857 BIG | cI857 RBS |
| Fwd Primer | 2 uL | 2 uL |
| Reverse Primer | 2 uL | 2 uL |
| PCR Supermix | 90 uL | 90 uL |
| Template | 2 uL | 2 uL |
| Water | 4 uL | 4 uL |
| Total | 100 uL | 100 uL |
Gel of PCR 8/1
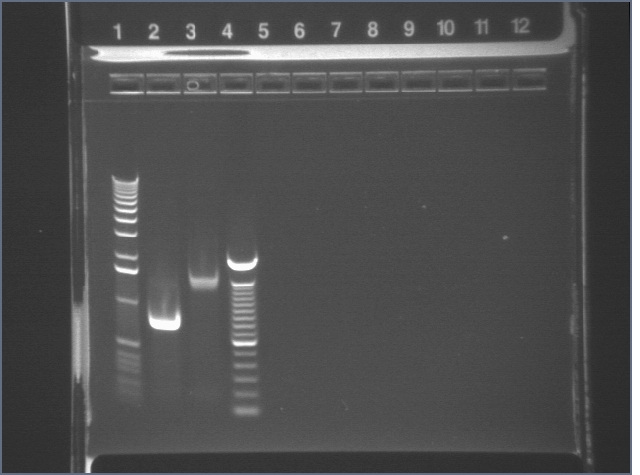
| 1% Agarose, visualized using EtBr/UV | |
|---|---|---|
| Lane | Sample | |
| 1 | 1 kb Ladder | |
| 2 | RBS | |
| 3 | BIG | |
| 4 | 100bp ladder | |
Used in TOPO TA Cloning Reaction and transformation with E1.
TOPO TA with Fresh cI PCR Product 8/1
| ' | RBS (0.5 uL DNA) | RBS (2 uL DNA) | RBS (4 uL DNA) | BIG (0.5 uL DNA) | BIG (2 uL DNA) | BIG (4 uL DNA) | Ctrl Template | Just Vector |
| PCR Product | 0.5 | 2 | 4 | 0.5 | 2 | 4 | 1 | 0 |
| Salt Solution | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| TOPO Vector | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Water | 3.5 | 2 | 0 | 3.5 | 2 | 0 | 3 | 4 |
Transformed in E1-- added either 2 or 4 uL of cloning reaction. Had limited and suspect X-gal so only applied to some plates, and included X-gal control for contamination.
Plate Results 8/2
Includes results for control reactions for TOPO TA cloning system. RBS indicates primer set RBS, and BIG indicated primer set BIG. These TOPO cloning reactions were done with fresh PCR product of cI857 (confirmed with E-gel). Volumes of DNA in parentheses indicate the amount of DNA added to the cloning reaction (0.5, 2, or 4 uL). Volumes of DNA rxn indicates how much of the cloning reaction was added to the competent cells (either 2 or 4 uL). Presence or absence of X-gal indicated in Description.
| Plate | Marker | # Colonies | Description |
| RBS (0.5 uL DNA) 2 uL rxn | Amp | 100+ | Had X Gal: 6 blue, rest white. All 1-2mm. |
| RBS (0.5 uL DNA) 4 uL rxn | Amp | 200+ | No X Gal: all white. All 1mm. |
| RBS (2 uL DNA) 2 uL rxn | Amp | 100+ | Had X Gal: ~25 blue, rest white. All 1-2 mm. |
| RBS (2 uL DNA) 4 uL rxn | Amp | 200+ | No X Gal: all white. All 0.5-1 mm. |
| RBS (4 uL DNA) 2 uL rxn | Amp | 100-200 | No X Gal: all white. All 0.5-1 mm. |
| RBS (4 uL DNA) 4 uL rxn | Amp | 100-200 big, ~5-10 tiny colonies | Had X-Gal: 40-50 blue, rest white. Bigger colonies 0.5-1mm, tiny colonies 0.1-0.5mm, flanking big colonies. |
| BIG (0.5 uL DNA) 2 uL rxn | Amp | ~75 | Had X-Gal: 23 blue, rest white. 0.5-1mm. |
| BIG (0.5 uL DNA) 4 uL rxn | Amp | 100+ | No X Gal: all white. All 1mm. |
| BIG (2 uL DNA) 2 uL rxn | Amp | 100-200 | No X-Gal: all white. All 0.5-1mm. |
| BIG (2 uL DNA) 4 uL rxn | Amp | 200+ | Had X-gal:50-100 blue, rest white. All 0.5mm. Some are tinged blue. |
| BIG (4 uL DNA) 2 uL rxn | Amp | 100 | No X-gal: All white, 1-2mm. |
| BIG (4 uL DNA) 4 uL rxn | Amp | 200 big, 25-50 tiny. | Had X-Gal: 40-50 blue, rest white. Bigger colonies 0.5-1mm, tiny colonies 0.1-0.5mm, flanking big colonies. |
| Ctrl Template 2 uL rxn | Amp | 75-100 | No X Gal: all white. All 0.5-1 mm. |
| Ctrl Template 4 uL rxn | Amp | 100 big, 25-50 tiny | Had X-Gal: 25-50 blue, rest white. Bigger colonies 0.5-1mm, tiny colonies 0.1-0.5mm, flanking big colonies. |
| Just Vector 2 uL rxn | Amp | ~35 | No X Gal: all white. All 1-2 mm. |
| Just Vector 4 uL rxn | Amp | 50+ | Had X-Gal: 2 fully white--one on edge where X-gal probably not present, one closer to center. Rest are blue, but some, especially close to the edge, are much lighter blue. Would recommend not picking even colonies that are tinged blue and to try to not pick around blue colonies. |
| (+) puc 19 ctrl | Amp | ~100 | All white, 1-2mm, not very evenly distributed. |
| (-) just cells | Amp | 0 | |
| (-) just X-gal | Amp | 0 |
Western Blot to Test IPTG and Heat Induction of Lac Systems
Induction and Preparing Lysate 7/29
| Sample | Protein Concentration |
| E1 P41 30 | 3.538645418 |
| E1 P41 30+IPTG | 3.397609562 |
| E1 P41 40 | 5.171713147 |
| E1 P84 30 | 3.413944223 |
| E1 P84 30+IPTG | 3.468924303 |
| E1 P84 40 | 4.507171315 |
| E1 P85 30 | 3.565338645 |
| E1 P85 30+IPTG | 3.680478088 |
| E1 P85 40 | 4.986055777 |
| E1 P30 30 | 3.359760956 |
| E1 P30 30 + IPTG | 0.502390438 |
| E1 P30 40 | 4.819123506 |
| Nat Cells 30 | 4.076494024 |
| Nat Cells OLD iptg | 4.479681275 |
| Nat Cells NEW IPTG | 4.276494024 |
| S1 p59B + p13 30 | 7.051394422 |
| S1 p59B + p13 IPTG | 5.019123506 |
| S1 P13 + P59b 1 30 | 6.422709163 |
| S1 P13 + P59b 1 IPTG | 5.163346614 |
| S1 P13 + P59b 2 30 | 6.125896414 |
| S1 P13 + P59b 2 IPTG | 4.337848606 |
| S1 P27 + P59b 2 30 | 2.854581673 |
| S1 P27 + P59b 2 IPTG | 4.25059761 |
| S1 P59b 30 | 6.311553785 |
| S1 P59b IPTG | 5.702788845 |
| S1 P13 (-) 30 | 5.293227092 |
| S1 P13 (-) IPTG | 6.001195219 |
Transformations in S1
Transformed mtrB constitutive system into S1 7/28
Electroporated and transformed S1.
| Plate | Marker | # Colonies | Description |
| E1 P3+mtrB notBB A | Kan | 50 | Small, with pink centers. Also some clusters of smaller colonies-- didn\'t pick any of those. |
| E1 P3+mtrB notBB B | Kan | 50 | Small, with pink centers. Also some clusters of smaller colonies-- didn\'t pick any of those. |
| E2 P1+mtrB A | Kan | 50 | Small, with pink centers. Also some clusters of smaller colonies-- didn\'t pick any of those. |
| E2 P1+mtrB B | Kan | 50 | Small, with pink centers. Also some clusters of smaller colonies-- didn\'t pick any of those. |
| E2 P3+mtrB NotBB A | Kan | 50 | Small, with pink centers. Also some clusters of smaller colonies-- didn\'t pick any of those. |
| E2 P3+mtrB NotBB B | Kan | 50 | Small, with pink centers. Also some clusters of smaller colonies-- didn\'t pick any of those. |
| (+) Ctrl (P59b) | Kan | 100s | Small, with pink centers. Also some clusters of smaller colonies. |
| (-) Ctrl (EB Buffer) | Kan | 0 |
Restreaked plates and picked two colonies per plate for liquid cultures and glycerol stocks.
Housekeeping
Made S1 Electrocompetent Cells 7/29
Made ~250 aliquots of wildtype and mtrB- Shewie-- 80 uL in each aliquot.
RE digests 07/29
We digested several samples of P17 in the P1/P3 vector with EX. We will add the high/low constitutive promoters.
Lane 1: 1 kb ladder
Lane 2: uncut E2 P3+17 B (3652)
Lane 3: E1 P1+17 A cut EX (3652)
Lane 4: E1 P1+17 B cut EX (3652)
Lane 5: E1 P1+17 C cut EX (3652)
Lane 6: E1 P1+17 D cut EX (3652)
Lane 7: E2 P1+17 A cut EX (3652)
Lane 8: E2 P1+17 B cut EX (3652)
Lane 9: E2 P3+17 A cut EX (3652)
Lane 10: E2 P3+17 B cut EX (3652)
Ligation with promoters
We attempted ligations of P38 and P39 with the proper sized P17s excised from the gels in a 2:6 ratio with TOP10 cells. No plates (Kan) had colonies.
Mutagenesis of Cph1 07/29
We set up a reaction to remove the PstI site from cph1 (P87A and B) according to the QuikChange kit protocol. We ran a PCR positive control and a transformation positive control. We transformed all of the samples in XL 1-Blue cells and P87B in TOP10 as well.
Transformation results 07/30
We used the TOP10 transformation protocol for both cell types. We plated 125 μL of each sample on two plates.
| Strain | Plate | DNA | # colonies (plate1, plate2) | Amount of DNA (μL) |
| E2 | Cm | 87A mut | ~400 each | 4 |
| E2 | Cm | 87B mut | TMTC, TMTC | 4 |
| E5 | Carb | PCR + control from kit | 160, 128 | 1 |
| E5 | Carb | pUC18 | TMTC, TMTC | 1 |
Ligations of digested 7/26 minipreps
See last 2 gels of last week's notebook.
All cells are of E1 and were on Kan plates.
| DNA | Vector:Insert | # Colonies |
| P3A+(E1 P38+51) | 1:7 | 1 |
| P3A+(E1 P38+51) | 1:5 | 1 |
| P3A+(E1 P38+51) | 2:6 | 2 |
| P3A+(E1 P39+51 A) | 1:7 | 17 |
| P3A+(E1 P39+51 A) | 1:5 | 5 |
| P3A+(E1 P39+51 A) | 2:6 | 0 |
| P3A+(E1 P39+51 C) | 1:7 | 1 |
| P3A+(E1 P39+51 C) | 1:5 | 0 |
| P3A+(E1 P39+51 C) | 2:6 | 0 |
1-2 non-fluorescent colonies were picked for each sample with colonies.
P3A+(E1 P39+51 C) didn't grow in liquid culture.
RE digests 07/30
Lane 1: 1 kb ladder
Lane 2: P3A + (E1 P38 +51 D) uncut (3155)
Lane 3: P3A + (E1 P38 +51 D) cut SP (3155)
Lane 4: P3A + (E1 P38 +51 C) cut SP (3155)
Lane 5: P3A + (E1 P38 +51 B) cut SP (3155)
Lane 6: P3A + (E1 P39 +51 A) uncut (3155)
Lane 7: P3A + (E1 P39 +51 A) cut SP (3155)
Lane 8: P3A + (E1 P39 +51 B) cut SP (3155)
Lane 9: P3A + (E1 P39 +51 C) cut SP (3155)
Lane 10: P3A + (E1 P39 +51 D) cut SP (3155)
Lane 11: E1 P63+98 B uncut
Lane 12: E1 P63+98 B cut XP (~2200)
Lane 13: E1 P63+98 A cut XP (~2200)
Lane 14: E2 P63+98 B cut XP (~2200)
Lane 15: E2 P63+98 A cut XP (~2200)
.
RE digests 07/31
gel 1
Lane 1: 1 kb ladder
Lane 2: E5P105D cut ApaLI and PstI
Lane 3: E2P105A cut ApaLI and PstI
Lane 4: E5P105F cut ApaLI and PstI
Lane 5: E5P105G cut ApaLI and PstI
Lane 6: E2P105E cut ApaLI and PstI
Lane 7: E5P105B cut ApaLI and PstI
Lane 8: E5P105H cut ApaLI and PstI
Lane 9: E2P105E cut ApaLI and PstI
Lane 10: E5P105A cut ApaLI and PstI
Lane 11: E2P105B cut ApaLI and PstI
Lane 12: E2P105B uncut
gel 2
Lane 2: 1 kb ladder
Lane 3: E2P105D uncut
Lane 4: E5P105C cut ApaLI and PstI
Lane 5: E2P105C cut ApaLI and PstI
Lane 6: E2P105E cut ApaLI and PstI
Lane 7: P87A uncut
Lane 8: P87A cut ApaLI and PstI
Lane 9: P87B cut ApaLI and PstI
Lane 10: E1P63+98 E uncut
Lane 11: E1P63+98 E cut XP
Lane 12: E2P63+98 E cut XP
Lane 13: E2P63+98 F cut XP
Lane 14: E2P63+98 F cut XP
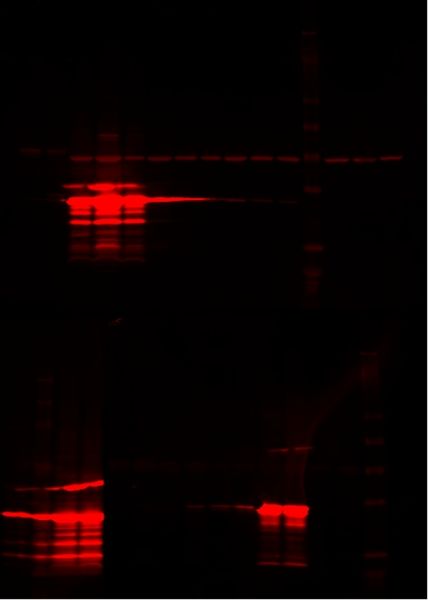
Western Blot
Testing ts-lac inducible system
Only the constitutive plasmids (p30 and p59b) and Natalie's plasmid when induced expressed the desired protein, indicating that neither the heat-inducible lac system nor the dual plasmid lac operon work.
Upper Gel: 1- S1 p13 and p59b 2 IPTG 2- S1 p13 and p59b 2 30°C 3- E1 p30 40°C 4- E1 p30 IPTG 5- E1 p30 30°C 6- E1 p85 40°C 7- E1 p85 IPTG 8- E1 p85 30°C 9- E1 p84 40°C 10- E1 p84 IPTG 11- E1 p84 30°C 12- Ladder 13- E1 p41 40°C 14- E1 p41 IPTG 15- E1 p41 30°C
Lower Gel: 1- pGEX (Natalie's cells) 30°C 2- Ladder 3- pGEX our IPTG 4- pGEX Silver Lab's IPTG 5- S1 p59b and p13 6- S1 p59b and p13 IPTG 7- S1 p13 and p59b 1 8- S1 p13 and p59b 1 IPTG 9- S1 p27 and p59b 10- S1 p27 and p59b IPTG 11- S1 p59b 12- S1 p59b IPTG 13- S1 p13 14- S1 p13 IPTG 15- Ladder
RE digests and ligations 08/02
Lane 1: 1 kb ladder
Lane 2: P3 cut XP, dephosphorylated (2750)
Lane 3: P39+51 cut XP (1405)
Gel extraction:
These bands were cut from the gel and melted at 65 °C. This was used in one set of ligations. Another set was done using our normal gel extraction protocol. We used ligation ratios of 2:6 and 3:5 of vector to insert. The ligations were transformed into DH5α cells.
Results
Both transformations involving the DNA extracted using the new gel extraction protocol failed (no colonies). The the ligation with a ratio of 2:6 of P3 to P39+51 did not give any colonies, but the 3:5 ligation plate had 2 colonies. Both of these colonies were picked and grown up in liquid cultures.
Cutting GFP out of P1
| Plate | Marker | # of Colonies | Description |
| tctr1 pUC19 (control) | Carb | ~125 | scattered 1mm diameter colonies |
| P1 A (XS) 10 min | Kan | Too many to count | ~0.5mm diameter colonies |
| P1 A (XS) 20 min | Kan | Too many to count | ~0.5mm diameter colonies |
| P1 A (XS) 40 min | Kan | Too many to count | ~0.5mm diameter colonies |
ON RE digests and ligations 08/03
- We cut P98 with ES to ligate it with a terminator, P63 cut with EX. This will give us RBS+mtrB+terminator. We also cut P98 with XmnI because the backbone and insert of P98 are around the same size, and digesting with XmnI cuts the backbone so it becomes smaller.
- We also cut the QPIs (P17: Tet and P51: Lac) in a p15A vector with EX. We will add high and low constitutive promoters.
Lane 1: 1 kb ladder
Lane 2: P98A uncut (~4200)
Lane 3: P98A cut ESXmnI (~2102; 1623, 433)
Lane 4: P98B cut ESXmnI (~2102; 1623, 433)
Lane 5: P98C cut ESXmnI (~2102; 1623, 433)
Lane 6: P98D cut ESXmnI (~2102; 1623, 433)
Lane 7: P98E cut ESXmnI (~2102; 1623, 433)
Lane 8: P98F cut ESXmnI (~2102; 1623, 433)
Lane 9: P63 cut EX (3284)
Lane 10: P3+51# cut EX (4120)
Lane 11: P1/3+51 cut EX (4120)
Lane 12: P3+51 cut EX (4120)
Lane 13: P3+17B uncut (3652)
Lane 14: P3+17 cut EX (~3652)
Lane 15: P3+17 cut EX (~3652)
Lane 16: 1 kb plus ladder
- Since the P63 band in lane 9 looked like it could have also had uncut DNA, we used a sample of P63 from 07/16 in the ligations.
- We used the standard QIAGEN gel extraction protocol.
- We ligated using 2:6 of vector to insert. We let the P98+63 ligations run for 10, 20 and 30 minutes (the other ligations only ran for 10 minutes).
- We transformed the P39+51, P39+17, P98+63 10' ligations into TOP10 cells. All of the other ligations were transformed into DH5α. We also transformed a positive control with pUC19.
- Since there was no SOC medium left, we incubated in LB for 2 hours after heat shock.
Results
| Plasmid | Ligation time | Number of colonies | Cell type |
| pUC19 control | N/A | 221 | DH5-alpha |
| P38+(P51 in P3) | 10' | 1 | TOP10 |
| P38+(P51 in P3) | 20' | 0 | DH5-alpha |
| P38+(P51 in P3) | 30' | 5 | DH5-alpha |
| P39+(P51 in P3) | 10' | 0 | TOP10 |
| P39+(P51 in P3) | 20' | 0 | DH5-alpha |
| P39+(P51 in P3) | 30' | 1 | DH5-alpha |
| P38+(P17 in P3) | 10' | 23 | TOP10 |
| P38+(P17 in P3) | 20' | 0 | DH5-alpha |
| P38+(P17 in P3) | 30' | 0 | DH5-alpha |
| P39+(P17 in P3) | 10' | 128 | TOP10 |
| P39+(P17 in P3) | 20' | 0 | DH5-alpha |
| P39+(P17 in P3) | 30' | 0 | DH5-alpha |
| P98+63 | 10' | TMTC (most) | TOP10 |
| P98+63 | 20' | TMTC | DH5-alpha |
| P98+63 | 30' | TMTC (least) | DH5-alpha |
 "
"