Team:Harvard/Dailybook/Week7/Chemical and Light
From 2008.igem.org
PCR
8/4: Cph/EnvZ, P1, P3, mtrB not BB
HO-pcyA and ompR bands excised, purified, and digested (ApaLI & Kpn overnight).
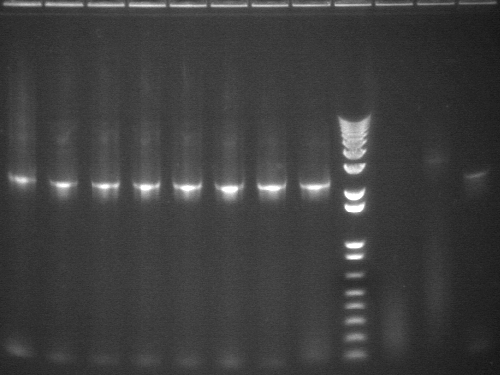
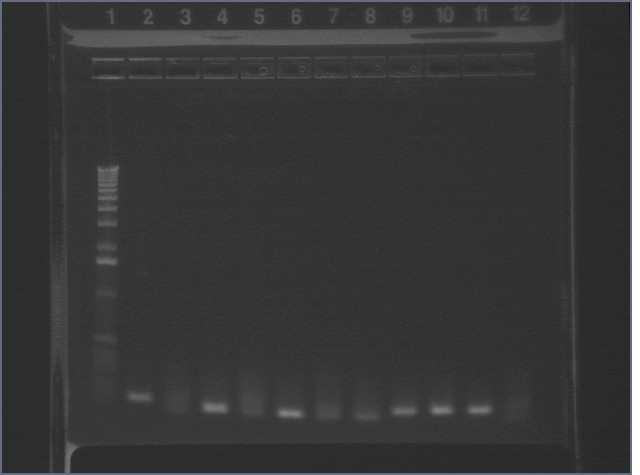
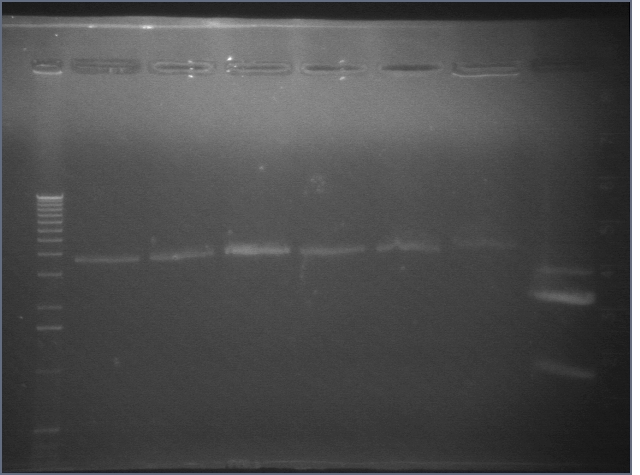
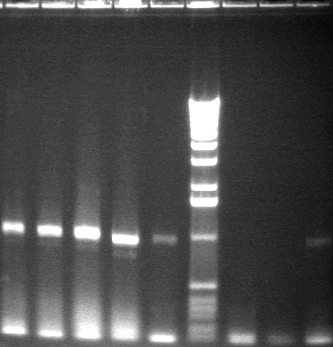
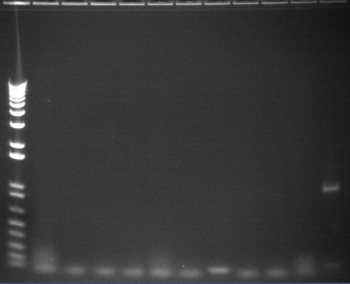
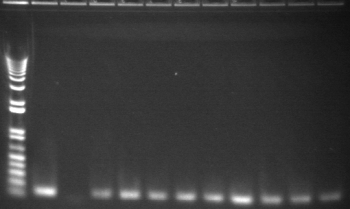
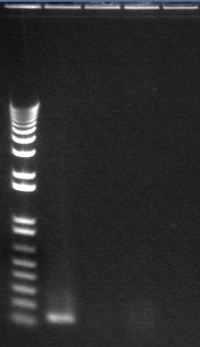
8/4 Colony PCR of putative mtrB samples
We picked 24 colonies each of old putative P98 and newly transformed putative P98+63 for colony PCR using the BioBrick primers and 55°C annealing for 30 cycles. If the colonies contained mtrB, a band at ~2.1 kb should appear. No colonies appeared to contain mtrB.
All gels with 1kb plus ladder
| P98 | P98 | P98+63 |
 |  | 
|
| P98+63 | Mix | |
 |  |
Lane 1: 1 kb plus ladder |
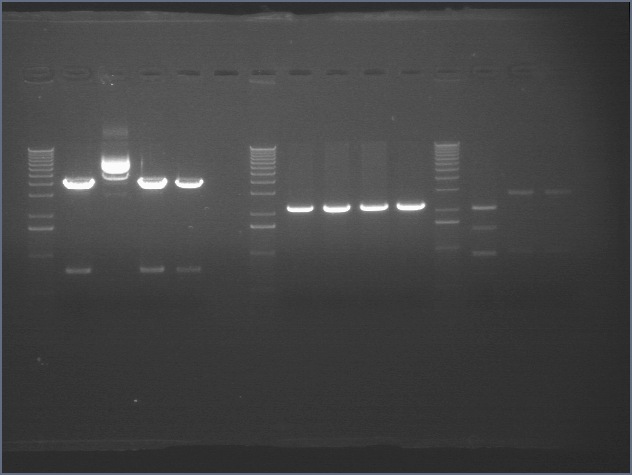
8/4 CphEnvZ, P1, P3, mtrB
Pl05 (CphEnvZ w/ PstI site mutated out) conditions: 5min @ 94°C → 40x [30s @ 94°C → 30s @ (50.8, 51.5, 52.6, 53.9, 55.4, 56.7, 57.7, 58.3)°C → 2:30 @ 72°C] → 7min @ 72°C → ∞ @ 4°C
8/5: gels
Bands excised and digested: Cph/P105 (AK), P3 (AK), mtrB (XP, ES)
Bands excised and saved: ompR, HO-pcyA, CDF
Ligations and transformations 08/06
We ligated P105 and P1/P3 not BB, and mtrB not BB and P1/P3 not BB. We tried letting the ligation reactions run for 5 and 10 minutes. We used P1/P3 not BB from 08/05 and from another date (older). We transformed using TOP10 and DH5α. We also retransformed P108, the Lac QPI with a high constitutive promoter in a p15A vector.
TOPO cloning
08/04
P1, mtrB
These had a few white colonies. Colony PCR, however, indicated that the correct PCR product was not incorporated.
ompR
All colonies were blue.
HO-pcyA
No colonies.
Transforming in XL10-Gold cells
| Plate | Marker | # Colonies | Description |
| XL10-Gold P75a + P63b (1:1) | Kan | 2 | No fluor |
| XL10-Gold P75b + P63b (1:1) | Kan | 6 | 1 fluor |
| XL10-Gold P75a + P63b (6:2) | Kan | 0 | |
| XL10-Gold P75b + P63b (6:2) | Kan | 1 | fluor |
| XL10-Gold P75a + P63b (7:1) | Kan | 1 | no fluor |
| XL10-Gold P75b + P63b (7:1) | Kan | 2 | both fluor |
| XL10-Gold Dephos P75a | Kan | 1 | |
| XL10-Gold Dephos p75b | Kan | 0 | |
| XL10-Gold P3 + (P39+p51) 2/6 | Kan | ||
| XL10-Gold P3 Topo w/ XGAL | Amp | ||
| XL10-Gold P1 Topo w/ XGAL | Amp | ||
| XL10-Gold mtrB Topo w/ XGAL | Amp | ||
| XL10-Gold pUC18 | Amp | 100s | White, very small |
| XL10-Gold negative control | Amp | many | extremely small |
| XL10-Gold negative control | Kan | 0 |
Ligations
HO-pcyA, ompR, CDF
All three ligations yielded >50 colonies on the TOP10 cell plate with a 5 minute ligation in all combinations (HO-pcyA and ompR with P1 and old P3; CDF with P3). The P90 (CDF+P3) did not fluoresce. The DH5α cells did not have nearly as many (<5) and ligation times (10min, 30 min) did not appear to be significant.
Colony PCR 8-6
We picked at least 18 colonies from each ligation set, patched a master plate, inoculated liquid cultures, and performed 12.5 μL colony PCR reactions. The BBp/sfx primer pair was used, annealing temperature was 50°C for first 10 cycles and 54°C for rest of the cycles. 2 controls were performed: one using a picked P20 colony, and another with miniprepped P20 plasmid.
Digests, 1-8
We ran the digests to see if PCR mutations could have introduced an internal cut site. However, even with the small amount of purified digest we had left, we could see a properly sized band for each of P1, ompR, and HO-pcyA PCR products.
9-19

| 1.2% E-gel visualized using EtBr/UV | |
|---|---|---|
| Lane | Contents (approx. expected band size) | |
| 1 | 1 kb plus ladder | |
| 2-11 | HO-pcyA + P1 ligation colony PCR (1.6kb) | |
| 12 | HO-pcyA + P3 ligation colony PCR (1.6kb) | |
20-30
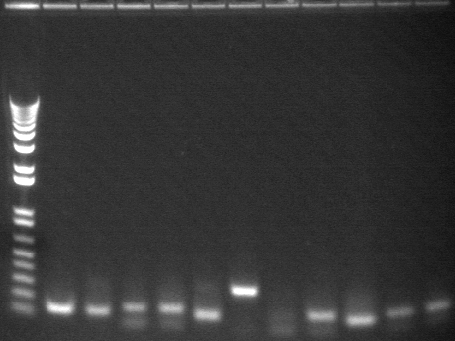
| 1.2% E-gel visualized using EtBr/UV | |
|---|---|---|
| Lane | Contents (approx. expected band size) | |
| 1 | 1 kb plus ladder | |
| 2-12 | HO-pcyA + P3 ligation colony PCR (1.6kb) | |
31-41

| 1.2% E-gel visualized using EtBr/UV | |
|---|---|---|
| Lane | Contents (approx. expected band size) | |
| 1 | 1 kb plus ladder | |
| 2-7 | HO-pcyA + P3 ligation colony PCR (1.6kb) | |
| 8-12 | CDF + P3 ligation colony PCR (900bp) | |
42-52
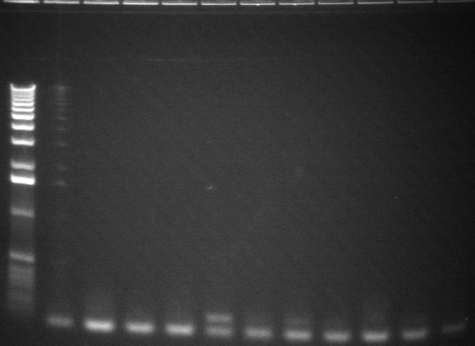
| 1.2% E-gel visualized using EtBr/UV | |
|---|---|---|
| Lane | Contents (approx. expected band size) | |
| 1 | 1 kb plus ladder | |
| 2-12 | CDF + P3 ligation colony PCR (900bp) | |
53-63

| 1.2% E-gel visualized using EtBr/UV | |
|---|---|---|
| Lane | Contents (approx. expected band size) | |
| 1 | 1 kb plus ladder | |
| 2-7 | CDF + P3 ligation colony PCR (900bp) | |
| 8-12 | ompR + P1 ligation colony PCR (1kb) | |
64-74

| 1.2% E-gel visualized using EtBr/UV | |
|---|---|---|
| Lane | Contents (approx. expected band size) | |
| 1-11 | ompR + P1 ligation colony PCR (1kb) (#64 and 73 contain ompR, as verified by sequencing) | |
| 12 | 1 kb plus ladder | |
75-85

| 1.2% E-gel visualized using EtBr/UV | |
|---|---|---|
| Lane | Contents (approx. expected band size) | |
| 1-2 | ompR + P1 ligation colony PCR (1kb) | |
| 3-11 | ompR + P3 ligation colony PCR (1kb) | |
| 12 | 1 kb plus ladder | |
86-94, P20 controls

| 1.2% E-gel visualized using EtBr/UV | |
|---|---|---|
| Lane | Contents (approx. expected band size) | |
| 1-9 | ompR + P3 ligation colony PCR (1kb) | |
| 10 | P20 colony PCR (1122) | |
| 11 | P20 plasmid PCR | |
| 12 | 1 kb plus ladder | |
Making Thermoinducible cI System
Ligation from 8/1
Plate Results (also listed in Week 6):
| Plate | Marker | # Colonies | Description |
| E1 P75a + P63b (1:1) | Kan | 0 | |
| E1 P75b + P63b (1:1) | Kan | 0 | |
| E1 P75a + P63b (6:2) | Kan | 1 | No Fluor, 3mm |
| E1 P75b + P63b (6:2) | Kan | 1 | Fluorescent, 1mm. |
| E1 P75a + P63b (7:1) | Kan | 0 | |
| E1 P75b + P63b (7:1) | Kan | 0 | |
| Dephos P75b | Kan | 0 |
Fluorescent colony was picked for liq culture and restreaked. Miniprep sent for sequencing-- ligation confirmed by sequencing (8/5).
Analytical Digest Gels from TOPO Cloning 8/5
Strange bands, but sent several clones for sequencing anyway.
Digestion and Ligation of P18+P63 and P75+P63
Done in tandem and side by side with Christina (except for digestion).
Digestion of P18, P63, and P75 8/4
| ' | P63 | P75b | P18 |
| DNA | 30 uL | 6 uL | 11 uL |
| 100x BSA | 0.5 uL | 0.25 uL | 0.25 uL |
| 10x Buffer | 5 uL Buffer 2 | 2.5 uL Buffer 2 | 2.5 uL Buffer 2 |
| RE 1 | EcoRI 1 uL | EcoRI 1 uL | EcoRI 1 uL |
| RE 2 | SpeI 1 uL | XbaI 1 uL | XbaI 1 uL |
| Water | 12.5 uL | 14.25 uL | 9.25 uL |
| Volume | 50 uL | 25 uL | 25 uL |
Gel of Digestion 8/5
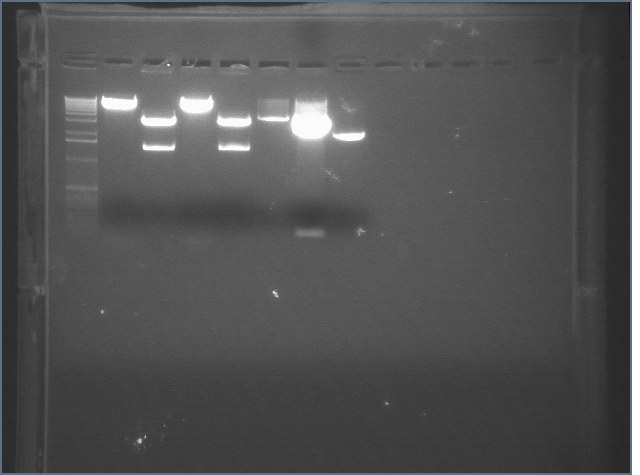
| 1% agarose gel visualized using EtBr/UV | |
|---|---|---|
| Lane | Sample | |
| 1 | 1 kB Ladder | |
| 2 | Christina\'s Vector (CE) | |
| 3 | Christina\'s Insert (CI) | |
| 4 | Christina\'s Vector (IE) | |
| 5 | Christina\'s Insert (IE) | |
| 6 | P75 EX | |
| 7 | P63 ES | |
| 8 | P18 EX | |
Ligation of P18+P63 and P75+P63 8/5
Used dephosphorylated and undephosphorylated vector.
Plate Results 8/6
| Plate | # Colonies | Notes |
| pUC19 Amp I | 31 | |
| Negative controls: | . | . |
| AmpI just cells | 0 | |
| AmpI ligC dephos. IE vector only | 8 | |
| AmpI ligC Ephos IE vector only v+i | 1 | |
| AmpI ligC G8 SP just vector | 6 | |
| AmpC ligC G8 SP just vector | 7 | |
| AmpC ligC dephos IE vector only | 5 | |
| DNA Positive controls: | . | |
| AmpC ligC G8 + G10 | >200 | |
| AmpI ligC G8 + G10 | 81 | |
| AmpI ligI G8 + G10 | 78 | |
| AmpC ligI G8 + G10 | 127 | |
| whole thing: | . | |
| AmpI ligC CE CG | 103 | |
| AmpI ligC CE dephos CG | 45 | |
| AmpC ligC CE dephos | 57 | |
| AmpC ligC CE CG | 224 | |
| uh oh: | . | |
| AmpI ligC IE CG | 3 | |
| AmpC ligC IE CG | 7 | |
| AmpC ligC IE dephos V + I | 46 | |
| iGEM DNA | . | |
| AmpI (-) | 3 | |
| AmpI (- Dephos) | 0 | |
| Kan 63+75 | 64 | |
| Kan 63 + 75 Dephos | 0 | |
| AmpI 63+ 18 | 100+ | |
| AmpI 63+18 dephos | 40 |
Digestion and Ligation of P63 + cI857 (Both Primer Sets)
Digestion of cI857 PCR product and P63 8/5
| ' | P63 EX | cI857 RBS (GP) ES | cI857 BIG (GP) ES | cI857 RBS (PP) ES | cI857 BIG (PP) ES | P104 EX |
| DNA | 10 uL | 3 uL | 7 uL | 10 uL | 10 uL | 10 uL |
| 100X BSA | 0.25 uL | 0.25 uL | 0.25 uL | 0.25 uL | 0.25 uL | 0.25 uL |
| 10X Buffer 2 | 2.5 uL | 2.5 uL | 2.5 uL | 2.5 uL | 2.5 uL | 2.5 uL |
| EcoRI | 1 uL | 1 uL | 1 uL | 1 uL | 1 uL | 1 uL |
| Enzyme 2 | 1 uL XbaI | 1 uL SpeI | 1 uL SpeI | 1 uL SpeI | 1 uL SpeI | 1 uL XbaI |
| Water | 10.25 uL | 17.25 uL | 13.25 uL | 10.25 uL | 10.25 uL | 10.25 uL |
| Volume | 25 uL | 25 uL | 25 uL | 25 uL | 25 uL | 25 uL |
Christina digested the same samples of P63, cI857 RBS GP and PP, and cI857 BIG GP and PP with EcoRI buffer. P104 was cut 8/6 in the morning.
Gel results 8/6

| 1% agarose gel visualized using EtBr/UV | |
|---|---|---|
| Lane | Sample | |
| 1 | 1 kB Ladder | |
| 2 | P63 | |
| 3 | P63c | |
| 4 | P104 | |
| 5 | 1 kB Ladder | |
Ligation of P63, P63c, and P104 with cI857 8/6
Accidentally mixed P63c and P104 so plated each of those ligations in Amp and Kan selective plates.
Plate results 8/7
| Plate | Final Insert Size | Marker | # Colonies |
| E1 P63 + RBS PP | 770 | Amp | 28 big |
| E1 P63 + RBS GPC | 770 | Amp | 25 big, 25 small |
| E1 P63 + BIG PP | 1695 | Amp | 50 big, 50 small |
| E1 P63 + BIG GPC | 1695 | Amp | 25 big, 25 small |
| E1 P63c + RBS PP | 770 | Amp | 50 big, 50 small |
| E1 P63c + RBS GPC | 770 | Amp | 25 big, 25 small |
| E1 P63c +BIG PP | 1695 | Amp | 25 big, 50 small |
| E1 P63c + BIG GPC | 1695 | Amp | 10 big, 25 small |
| E1 P63 + RBS PP 6:2 | 770 | Amp | ~50 |
| E1 P63 + RBS PP 4:4 | 770 | Amp | 11 |
| E1 P63 + BIG PP (No dephos) | 1695 or 1600 | Amp | 0 |
| E1 P63 Dephos only | N/a | Amp | 0 |
| E1 P63c Dephos only | N/a | Amp | 10 big, 25 small |
| E1 P104 No dephos | 1020 | Amp | 0 |
| E1 P104 Dephos only | n/a | Amp | 75-100 |
| E1 P104 + RBS GP | 1695 | Amp | 50 |
| E1 P104 + RBS PP | 1695 | Amp | 50 |
| E1 P104 + BIG GP | 2620 | Amp | 0 |
| E1 P104 + BIG PP | 2620 | Amp | 0 |
| E1 P104 + RBS GPC | 1695 | Amp | 3 |
| E1 P104 + RBS PPC | 1695 | Amp | 5 |
| E1 P104 + BIG GPC | 2620 | Amp | 0 |
| E1 P104 + BIG PPC | 2620 | Amp | 4 |
| puc 19 (+) ctrl | N/a | Amp | 50-75 |
| (-) ctrl just cells | N/a | Kan | 0 |
| E1 P63c + RBS PP | 770 | Kan | ~20 |
| E1 P63c + RBS GPC | 770 | Kan | 3 big (2mm) |
| E1 P63c +BIG PP | 1695 | Kan | 0 |
| E1 P63c + BIG GPC | 1695 | Kan | 0 |
| E1 P63c Dephos only | N/a | Kan | 0 |
| E1 P104 No dephos | 1020 | Kan | 0 |
| E1 P104 Dephos only | n/a | Kan | 0 |
| E1 P104 + RBS GP | 1695 | Kan | 0 |
| E1 P104 + RBS PP | 1695 | Kan | 16 big (2mm) |
| E1 P104 + BIG GP | 2620 | Kan | 0 |
| E1 P104 + BIG PP | 2620 | Kan | 0 |
| E1 P104 + RBS GPC | 1695 | Kan | 1 |
| E1 P104 + RBS PPC | 1695 | Kan | 0 |
| E1 P104 + BIG GPC | 2620 | Kan | 0 |
| E1 P104 + BIG PPC | 2620 | Kan | 0 |
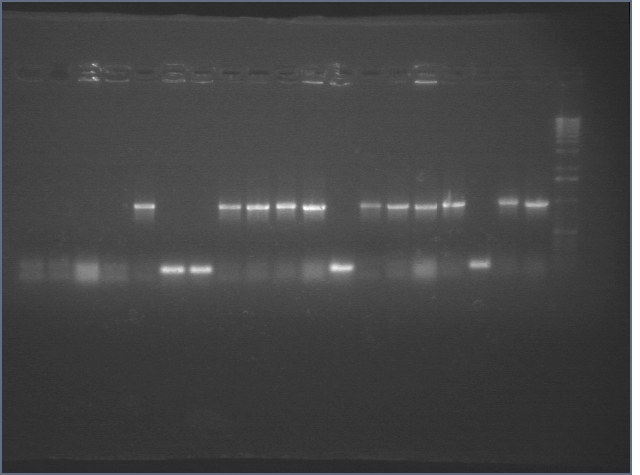
Colony PCRs of 8/6 Ligations 8/7
E1 P63 + RBS PP B to be sequenced Monday. All of the bands in the third gel indicated re-ligated vector.
Ligation of P63, P63c, and P104 with cI (repeat) 8/7
Ligations were re-done with newly digested vector because of mishap in 8/6 ligations.
Digestions of P63, P63c, and P104 8/6
| ' | P63 EX | P63c EX | P104 EX |
| DNA | 10 uL | 10 uL | 10 uL |
| 100X BSA | 0.25 uL | 0.25 uL | 0.25 uL |
| 10X Buffer | 2.5 uL of Buffer 2 | 2.5 uL of EcoRI Buffer | 2.5 uL of Buffer 2 |
| EcoRI | 1 uL | 1 uL | 1 uL |
| Enzyme 2 | 1 uL XbaI | 1 uL XbaI | 1 uL XbaI |
| Water | 10.25 uL | 10.25 uL | 10.25 uL |
| Volume | 25 uL | 25 uL | 25 uL |
Gel of Digestion 8/7

| 1% agarose gel visualized using EtBr/UV | |
|---|---|---|
| Lane | Sample | |
| 1 | 1 kB Ladder | |
| 2 | P63 | |
| 3 | P63c | |
| 4 | P104 | |
| 5 | 1 kB Ladder | |
Bands were extracted, purified, and dephosphorylated.
Ligation of P63, P63c, and P104 with cI 857 8/7
Vector backbones were ligated with cI857 digested 8/5.
Plate Results 8/8
| Plate | Final Insert Size | Marker | # Colonies (After 1 Day) | # Colonies After 1 Day |
| E1 P63 (Buffer 2) + RBS GP | Amp | 0 | N/a | |
| E1 P63 (Buffer 2) + BIG PPC | Amp | 5 1-1.5mm colonies | N/a | |
| E1 P63c + RBS GP | Amp | 1 1.5mm colony | N/a | |
| E1 P63c +BIG PPC | Amp | 6 1-1.5mm colonies | N/a | |
| E1 P63 (2) Dephos only | Amp | 2 1mm colonies on edge | N/a | |
| E1 P63c Dephos only | Amp | 1 1.5mm colony | N/a | |
| E1 P104 + RBS GP 6:2 | Kan | 3 colonies, no fluor | 3 2-3mm colonies, 6 1mm colonies | |
| E1 P104 + RBS GP | Kan | 8 colonies, no fluor | 8 2-3mm colonies, 3 1mm colonies | |
| E1 P104 + RBS PP | Kan | 30 colonies, 10 fluor, rest no fluor | 30 big 2-3mm colonies, 2 1mm colonies. | |
| E1 P104 + BIG GP | Kan | 4 small 0.25 colonies? | 4 1mm colonies. | |
| E1 P104 + BIG PP | Kan | 0 colonies | 5 small 0.25mm colonies. | |
| E1 P104 + RBS GPC | Kan | 3 1.5mm colonies, no fluor | 3 big 3mm colonies, 2 1mm colonies. | |
| E1 P104 + RBS PPC | Kan | 1 0.5mm colony | 1 2mm colony, no fluor, and 2 1mm colonies. | |
| E1 P104 + BIG GPC | Kan | Lots of small 0.25mm colonies | 100 0.5-1mm colonies, no fluor. | |
| E1 P104 + BIG PPC | Kan | 0 colonies | 1 0.5mm colony | |
| E1 P104 + BIG PPC 6:2 | Kan | 0 colonies | 0 | |
| E1 P104 Dephos only | Kan | 1 colony on edge of plate 1.5 mm | 2mm colony on edge, and 3 0.5-1mm colonies. | |
| puc 19 (+) ctrl | Amp | 100-200 1.5mm colonies | N/a | |
| Kan (-) ctrl | Kan | 0 | 1 1mm colony | |
| Amp (-) ctrl | Amp | 0 | N/a |
Restreaked some non-fluor colonies from RBS ligations/ transformations to see if repressor is being repressed-- grew in 37 and 42 degrees. No perceived difference in fluor/ no induction.
Colony PCR of Ligations 8/8
3 bands in second gel indicate re-ligated vector.
Ligation of P104 and P63+P18 with cI RBS from TOPO
Digestion of cI RBS from TOPO and P18+P63 8/8
P18+ P63 not confirmed by sequencing yet.
| ' | cI857 RBS TOPO ES 1 | cI857 RBS TOPO ES 2 | cI857 RBS TOPO ES 3 | cI857 RBS TOPO ES 5 | P18+ P63 ES 1 | P18+ P63 ES 2 | P18+ P63 ES 3 | P18+ P63 ES 4 | E1 P63 + RBS PP B | E1 P104 + RBS PP A | E1 P63c + RBS PP G |
| DNA | Fill in | Fill in | Fill in | Fill in | Fill in | Fill in | Fill in | Fill in | Fill in | Fill in | Fill in |
| 100X BSA | 0.25 uL | 0.25 uL | 0.25 uL | 0.25 uL | 0.25 uL | 0.25 uL | 0.25 uL | 0.25 uL | 0.25 uL | 0.25 uL | 0.25 uL |
| 10X Buffer 2 | 2.5 uL | 2.5 uL | 2.5 uL | 2.5 uL | 2.5 uL | 2.5 uL | 2.5 uL | 2.5 uL | 2.5 uL of Buffer 3 | 2.5 uL of Buffer 3 | 2.5 uL of Buffer 3 |
| EcoRI | 1 uL | 1 uL | 1 uL | 1 uL | 1 uL | 1 uL | 1 uL | 1 uL | 1 uL XbaI | 1 uL XbaI | 1 uL XbaI |
| Enzyme 2 | 1 uL SpeI | 1 uL SpeI | 1 uL SpeI | 1 uL SpeI | 1 uL XbaI | 1 uL XbaI | 1 uL XbaI | 1 uL XbaI | 1 uL PstI | 1 uL PstI | 1 uL PstI |
| Water | Fill in | Fill in | Fill in | Fill in | Fill in | Fill in | Fill in | Fill in | Fill in | Fill in | Fill in |
| Volume | 25 uL | 25 uL | 25 uL | 25 uL | 25 uL | 25 uL | 25 uL | 25 uL | 25 uL | 25 uL | 25 uL |
Gel results 8/9
RBS TOPO 1,3, and 5, and P18+ P63 EX 1-4 were extracted and purified.
Update: At least P18+ P63 1-2 have been sequence confirmed.
Digest of P104 and P63 8/9
| ' | P63 EX | P104 EX |
| DNA | 5 uL | 15 uL |
| 100X BSA | 0.25 uL | 0.25 uL |
| 10X Buffer | 2.5 uL of Buffer 2 | 2.5 uL of Buffer 2 |
| EcoRI | 1 uL | 1 uL |
| Enzyme 2 | 1 uL XbaI | 1 uL XbaI |
| Water | 15.25 uL | 5.25 uL |
| Volume | 25 uL | 25 uL |
Gel of Digestion 8/10
Extracted and purified 8/10.
mtrB, P97, P63 digests 08/06
We digested mtrB with ES and XP so that we can put it into the P63 (terminator) and P97 (RBS) vectors. We also digested P97 with SP (and dephosphorylated it).
Ligation transformation results 08/07
| Plasmid | Ligation time | Ligation ratio (vector:insert) | Strain | No. of Colonies |
| mtrB not BB+P1 not BB old | 10 min | 2:6 | DH5-alpha | 1 |
| mtrB not BB+P1 not BB | 10 min | 2:6 | DH5-alpha | 4 |
| mtrB not BB+P1 not BB | 5 min | 2:6 | TOP10 | 88 |
| mtrB not BB+P3 not BB old | 10 min | 2:6 | DH5-alpha | 0 |
| mtrB not BB+P3 not BB | 10 min | 2:6 | DH5-alpha | 8 |
| mtrB not BB+P3 not BB | 5 min | 2:6 | TOP10 | 40 |
| mtrB+P63 | 10 min | 2:6 | TOP10 | 48 |
| mtrB+P63 | 10 min | 1:7 | DH5-alpha | 5 |
| mtrB+P97 | 10 min | 2:6 | DH5-alpha | TMTC |
| mtrB+P97 | 10 min | 1:7 | DH5-alpha | 200 |
| P105+P1 not BB old | 10 min | 2:6 | DH5-alpha | 5 |
| P105+P1 not BB | 10 min | 2:6 | DH5-alpha | 1 |
| P105+P1 not BB | 5 min | 2:6 | TOP10 | 2 |
| P105+P3 not BB old | 10 min | 2:6 | DH5-alpha | 1 |
| P105+P3 not BB | 10 min | 2:6 | DH5-alpha | 8 |
| P105+P3 not BB | 5 min | 2:6 | TOP10 | 55 |
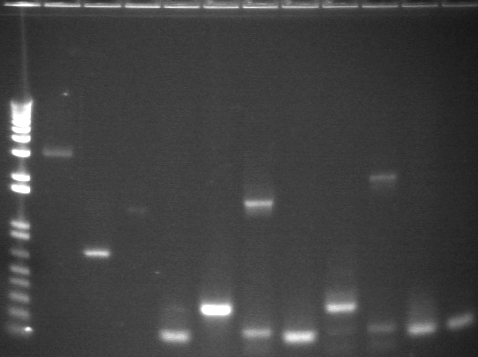
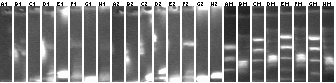
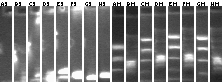
Colony PCR
Ladders (end of each block) alternate as low range (100, 200, 400, 800, 2000bp) and 1kb plus. Expected band sizes indicated. (Lanes rearranged from 2% agarose 96 well E-gel)
1-16
Lane 1: Plasmid control P85 (2.3kb)
Lane 2: Colony control P88 (2.2kb)
Lanes 3-16: P105+P3 ligation colony PCR (2.5kb)
4, 6, 14 look like they have potentially correct products, so liquid cultures were grown up.
14 is incorrect- it's just P1 in pSB3K3.
17-24: P105+P1 ligation colony PCR (2.5kb)
None appear to be correct.
25-48: mtrB+P63 ligation colony PCR (2.2kb)
27, 28 picked.
RE digests 08/09
Lane 1: 1 kb ladder
Lane 2: mtrB+63 #28 cut XP (~2200+3.1kb)
Lane 3: E1P3A-P38+51C cut SP (4155)
Lane 4: E1P3A-P38+51D cut SP (4155)
Lane 5: E1P3A-P38+51B cut SP (4155)
49-64: mtrB+P97 ligation colony PCR (2.1kb)
51 picked.
65-80: mtrB'+P3 ligation colony PCR (2.3kb)
65, 69, 70, 78, 79, 80 picked.
65 sequence did not read properly.
81-96: mtrB'+P1 ligation colony PCR (2.3kb)
82, 87, 95 picked.
82 is wrong- mystery sequence. 87 and 95 are BLAST to pHELLSGATE ?!
RE digests 08/07
Lane 1: 1 kb ladder
Lane 2: E1P1+P17A cut EX (3652)
Lane 3: E1P1+P17C cut EX (3652)
Lane 4: E1P1+P17D cut EX (3652)
Lane 5: E2P1+P17A cut EX (3652)
Lane 6: E2P1+P17B cut EX (3652)
Lane 7: P108 cut SP (4155)
Lane 8: P45 cut XP (876; 2079)
Ligations 08/08
- We ligated P17 in a p15A vector (P1 or P3) to P38 and P39. We used a vector to insert ratio of 1:7. We also prepared a ligation reaction with 1 μL vector (P17) and 7 μL water as a transformation control. We transformed 50 μL TOP10 cells with 7 μL DNA and 100 μL DH5α cells with 7 μL DNA. We also used 7 μL of the vector-only ligation to transform 7 μL DH5α cells.
- To test the Lac QPI system, we ligated P45 (RBS+GFP+terminator) to P108. We used a vector to insert ratio of 2:6. We also prepared a ligation reaction with 1 μL vector (P108) and 7 μL water as a transformation control. We transformed 100 μL DH5α cells with 7 μL DNA and we also used 7 μL of the vector-only ligation to transform 7 μL DH5α cells.
Transformation results 08/09
| Plasmid | Amount of DNA | Strain | Ligation ratio (vector:insert by volume) | Number of colonies |
| P108 (vector control) | 7 ul | 100 ul DH5-alpha | 2 (vector)!6 (water) | 0 |
| P45+108 | 7 ul | 100 ul DH5-alpha | 2:6 | 1 |
| P17 in p15A (vector control) | 7 ul | 100 ul DH5-alpha | 1 (vector):7 (water) | 188 |
| (P17 in p15A)+P39 | 7 ul | 50 ul TOP10 | 1:7 | 1 |
| (P17 in p15A)+P39 | 7 ul | 100 ul DH5-alpha | 1:7 | 0 |
| (P17 in p15A)+P38 | 7 ul | 50 ul TOP10 | 1:7 | 3 |
| (P17 in p15A)+P38 | 7 ul | 100 ul DH5-alpha | 1:7 | 0 |
Colony PCR 8/9
We picked all of the noncontrol colonies and one control colony for colony PCR using the BBp/sfx primers.
Since the bands are at least plausible, the P38/39 in P1/3 samples were grown up and miniprepped.
Retransformation 8/9
We retransformed 5μL of the P39+17 in P1/3 ligation into 50μL DH5α and obtained two colonies.
Colony PCR shows that one of these does not contain the right construct. The other was set up in 5ml culture.
1kb ladder, colony A, colony B (both should have ~950 products if correct)
New ligations and transformations 08/10
We retransformed our old P108+45 (2:6 vector to insert) ligation and the old vector control. We also did this ligation over with a 2:1 molecular ratio of insert to vector. Both P108 (4155 bp) and P45 (876 bp) were about 17 ng/μL. We used a ratio of 5.4:2.3 vector to insert by volume.
Transformation results
| Plasmid | Ratio (vector!insert) | Strain | Number of colonies |
| P108 vector control (old) | 2 ul P108!6 ul EB | DH5-alpha | 0 |
| P108+45 (old) | 2!6 | DH5-alpha | 0 |
| P108+45 (old) | 2!6 | TOP10 | 4 |
| P108 vector control (new) | 5.4 ul P108!2.3 ul EB | DH5-alpha | 0 |
| P108+45 (new) | 5.4 ul P108!2.3 ul EB | DH5-alpha | 2 |
| P108+45 (new) | 5.4 ul P108!2.3 ul EB | DH5-alpha | 8 |
Transforming LacZ parts
We attempted to transform the following parts: I732017, E0435, E0435, I732005 into our competent DH5α. E0435 (on CM) had 0 colonies). The others (on Carb) had 10-20 extremely small colonies (after 16+hrs of incubation) each. We picked 2 of each for liquid culture, but none grew.
We requested the parts directly from MIT.
 "
"