Team:USTC/Project
From 2008.igem.org
| Home | The Team | The Project | Components | Results | Parts Submitted to the Registry | Notebook |
|---|
Contents |
Abstract
It is an amazing process in nature that the evolution from Protozoa to Metazoa. Even in the development of each Metazoa, it is still unknown how the genome regulates stem cells to differentiate into different kinds of cells, which can compose different tissues or organs, according to where they are in the body. There should be a self-organized process. Here we are trying to build a self-organized multiple-cell system based on the quorum sensing system to understand the mechanism of this process. We employed small molecules in the AHL family as messengers to transmit the orders of differentiation, and we use Cre recombinase as the executor of differentiation. With the use of an artificially designed network, we are trying to construct a new kind of cells, through which a ring composed by GFP will be seen on the plate if the colony is big enough.
Introduction
Background
Self-organization is a process of attraction and repulsion in which the internal organization of a system, usually an open system, increases its complexity without being affected by the source outside. It is a widely observed phenomenon in nature, such as spontaneous folding of proteins and other bio-macromolecules, formation of bilayer lipid membranes, creation of caste by social animals, etc. Self-organization appears in different levels of living activities, from the formations of macromolecules to the activities of organisms. One of these phenomena that are paid close attention to is pattern formation, which is observed in the development of living organisms and also the evolution from Protozoa to Metazoa. There are two important processes in pattern formation: differentiation and organization. Many efforts were reported to reproduce differentiation by designing a specific gene regulatory network in E.coli to mimic the differentiation process in multi-cell organisms. Meanwhile, it has been attempted for many times to re-build the organization of multi-cell system. For instance, a ring formed by GFP has been achieved on the plate by manually locating together two different types of cells --“sender” cells and “receiver” cells. But the two processes are achieved respectively in E.coli.
In our perspective, the combination of the two processes to imitate the whole developing procedure from a single cell to a self-organized multi-cellular system can enable us to know more about the molecular mechanism of development, and even the evolutionary process. In view of this, our project is focused on how to integrate them together.
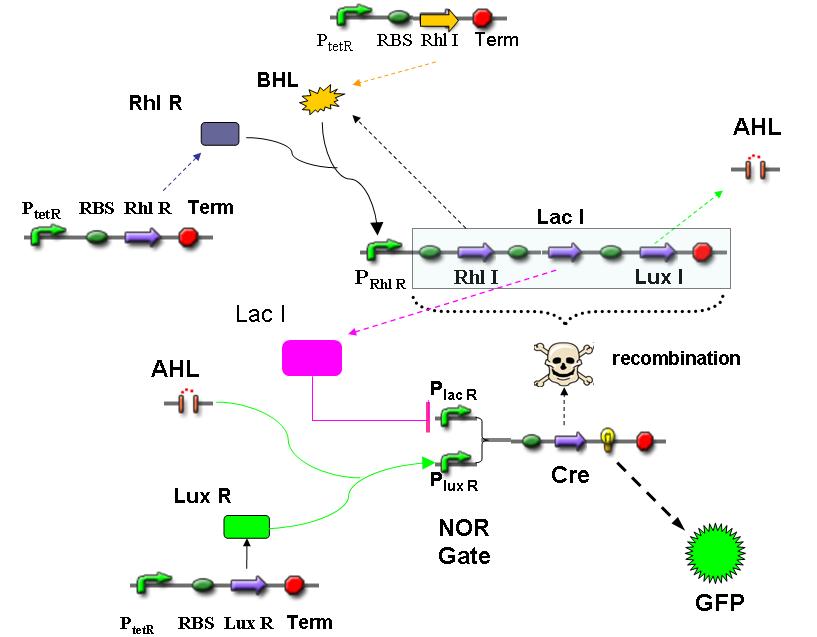
Design Gene Regulatory Network
We design a time-dependent gene regulatory network to construct a self-organized system. The RhlI/RhlR system is employed to report the cell density. The “sender” cell will appear first when the density reaches a certain level. And it will command other cells around it to differentiate into “receiver” cells by sending the signal molecules into the environment. After receiving the signals, the Cre recombinase in “receiver” cells will be generated and "scissor" the capacity of sending signals. In this way, a spatial differentiation will be accomplished, and the result will be that the “sender” cells are surrounded by the “receiver” cells. The chemical gradient of the signal molecules will be built up sometime after the differentiation process is finished. In this way, the response in a specific region will eventually result in a picture of a green ring in the black background.
It is just the beginning for self-organized graphics in bacteria. Now that the formed ring tells us how the bacteria respond to distance, in the next step, possibly we can show how the bacteria respond to angle. After that, we will have a better understanding about why we look as what we appear now.
 "
"