Team:Chiba/Calendar-Home/1 September 2008
From 2008.igem.org
(Difference between revisions)
(→Team:Input) |
(→Team:Input) |
||
| (One intermediate revision not shown) | |||
| Line 53: | Line 53: | ||
#Incubated cultures(from Glycerol Stocks) with 2mL of LB-Ampicillin Medium for 12 hours at 37 degrees. | #Incubated cultures(from Glycerol Stocks) with 2mL of LB-Ampicillin Medium for 12 hours at 37 degrees. | ||
| - | OD: | + | OD:UV⊕:5.21,UV⊖(negative control):5.38 |
| - | #moved the cultures to small plates and started UV-irradiation.(wavelength:254nm,distance from the UV lamp to the cultures were 7.5cm.Put the negartive control in | + | #moved the cultures to small plates and started UV-irradiation.(wavelength:254nm,distance from the UV lamp to the cultures were 7.5cm.Put the negartive control in a dark place. |
#covered the plates with polyethylene wrap. | #covered the plates with polyethylene wrap. | ||
#after irradiation(Sample:0min,10min,30min,1h,2h,4h,6h,8h)(Negative Control:0h,1h,4h,8h),we agitate the culture and took 20μl from each other | #after irradiation(Sample:0min,10min,30min,1h,2h,4h,6h,8h)(Negative Control:0h,1h,4h,8h),we agitate the culture and took 20μl from each other | ||
Latest revision as of 01:42, 30 October 2008
31 August 2008 <|> 2 September 2008
Contents |
Laboratory work
Team:Input
- Ptet+RBS+cI
Transformation(Double Transformation)
- -Ptet-cI-pMB1-Amp-,-PcI-GFP-p15A-Cm-(JW1908Δflic)
- -Ptet-cI-pMB1-Amp-
| Double | Single | |
| dH2O(μL) | 1 | 2 |
| XbaI(μL) | 1 | 1 |
| SpeI(μL) | 1 | - |
| BSA(×10)(μL) | 1 | 1 |
| NEB(×10)(μL) | 1 | 1 |
| DNA(μL) | 5 | 5 |
| TOTAL(μL) | 10 | 10 |
UV irradiation test
- two plasmids from (JW1908)
- -Ptrc-LuxR-Plux-cI-colE1-Amp-
- -PcI-GFP-p15a-Cm-
- Incubated cultures(from Glycerol Stocks) with 2mL of LB-Ampicillin Medium for 12 hours at 37 degrees.
OD:UV⊕:5.21,UV⊖(negative control):5.38
- moved the cultures to small plates and started UV-irradiation.(wavelength:254nm,distance from the UV lamp to the cultures were 7.5cm.Put the negartive control in a dark place.
- covered the plates with polyethylene wrap.
- after irradiation(Sample:0min,10min,30min,1h,2h,4h,6h,8h)(Negative Control:0h,1h,4h,8h),we agitate the culture and took 20μl from each other
- diluted each cultures with LB-ampicillin medium 104-fold and 105-fold (volume/volume).
- incoculated 20μl of the cultures and incubated for 12 hours at 37 degrees.
- counted the CFU(determined viable cell count).
Viable cell count
| UV+ 104 | UV+ 105 | UV-104 | UV-105 | |
| 0min | 423 | 74 | 271 | 156 |
| 10min | 301 | 51 | - | - |
| 30min | 139 | 7 | - | - |
| 1h | 89 | 5 | 1040 | 54 |
| 2h | 5 | 1 | - | - |
| 4h | 2 | 0 | 254 | 50 |
| 6h | 0 | 0 | - | - |
| 8h | 1 | 0 | 1155 | 106 |
Team:Communication
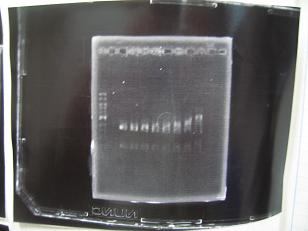
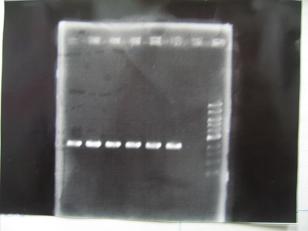
- (31/8)--->Gel Check

|
|
|
|
|
|
- --->Gel extract
- --->zymo
- insert-1([http://partsregistry.org/Part:BBa_I9026 BBa_I9026]) -> 7μL
- insert-2([http://partsregistry.org/Part:BBa_I9030 BBa_I9030]) -> 7μL
- insert-3([http://partsregistry.org/Part:BBa_S03154 BBa_S03154]) -> 7μL
- vector-4([http://partsregistry.org/Part:BBa_R0010 BBa_R0010]) -> 15μL
- --->SAP
- vector-4([http://partsregistry.org/Part:BBa_R0010 BBa_R0010])
- --->Zymo
- vector-4([http://partsregistry.org/Part:BBa_R0010 BBa_R0010]) -> 20μL
- --->Gel Check

|
|
- --->Ligation
| Sample No. | (1) | (2) | (3) | (4) | (5) |
| insert-1([http://partsregistry.org/Part:BBa_I9026 BBa_I9026]) | 3 | - | 3 | - | - |
| insert-2([http://partsregistry.org/Part:BBa_I9030 BBa_I9030]) | - | 3 | - | 3 | - |
| vector-4([http://partsregistry.org/Part:BBa_R0010 BBa_R0010]) | 3 | 3 | - | - | 3 |
| ligase | 1 | 1 | 1 | 1 | 1 |
| Buffer | 1 | 1 | 1 | 1 | 1 |
| dH2O | 2 | 2 | 5 | 5 | 5 |
| TOTAL | 10μl | 10μl | 10μl | 10μl | 10μl |
- --->Transformation
- Competent cells : XL10GOLD 30μL
- Transformed the following and grew on new ampicillin plates.
- [http://partsregistry.org/Part:BBa_K084009 BBa_K084009(Plac+RBS+RhlI+LVA, Amp)] -> 628 colonies
- [http://partsregistry.org/Part:BBa_K084010 BBa_K084010(Plac+RBS+CinI+LVA, Amp)] -> 500 colonies
- insert-1(RBS+RhlI+LVA) -> 9 colonies
- insert-2(RBS+CinI+LVA) -> No colonies on the plate
- vector-4(Plac, Amp) -> 186 colonies
- --->(2/9) Colony PCR
- Competent cells : JW1908 40μL
- Transformed the following and grew on new ampicillin plates.
- [http://partsregistry.org/Part:BBa_K084007 BBa_K084007(Plac+RBS+LasI)]
- [http://partsregistry.org/Part:BBa_K084008 BBa_K084008(Plac+RBS+RhlI)]
- [http://partsregistry.org/Part:BBa_K084010 BBa_T9002(Ptet+RBS+LuxR+GFP)]
- --->(2/9)Liquid Culture
Team:Output
- vector:[http://partsregistry.org/Part:BBa_R0010 BBa_R0010] insert:[http://partsregistry.org/Part:BBa_J52008 BBa_J52008](1)
- negative control:[http://partsregistry.org/Part:BBa_R0010 BBa_R0010](2)
| Sample No. | 1 | 2 |
| vector | 2 | 2 |
| insert | 3 | 0 |
| Ligase Buffer | 2 | 2 |
| Ligase | 1 | 1 |
| dH2O | 3 | 5 |
| TOTAL | 10μl | 10μl |
-->R/T 2hour
- [http://partsregistry.org/Part:BBa_R0079 BBa_R0079]
- [http://partsregistry.org/Part:BBa_R0071 BBa_R0071]
- [http://partsregistry.org/Part:BBa_R0077 BBa_R0077]
- [http://partsregistry.org/Part:BBa_R0078 BBa_R0078]
- [http://partsregistry.org/Part:BBa_R0062 BBa_R0062]
 "
"