DNA-Origami
From 2008.igem.org
(Difference between revisions)
WeberSimone (Talk | contribs) |
m |
||
| (18 intermediate revisions not shown) | |||
| Line 4: | Line 4: | ||
<font face="Arial Rounded MT Bold" style="color:#010369">_DNA-Origami</font></div> | <font face="Arial Rounded MT Bold" style="color:#010369">_DNA-Origami</font></div> | ||
<br><br> | <br><br> | ||
| - | + | <h2>Introduction</h2> | |
| + | [[Image:Freiburg2008_Fab_on_Origami_animated.gif|right|400 px]] | ||
Paul Rothemund has discovered that it is possible to shape M13-Phage single-strand-DNA simply adding oligonucleotides that will work as „brackets“ when complementing the long single-strand.<br> In this way, one can generate for example DNA-squares of a certain size with „nods“ at certain distances. | Paul Rothemund has discovered that it is possible to shape M13-Phage single-strand-DNA simply adding oligonucleotides that will work as „brackets“ when complementing the long single-strand.<br> In this way, one can generate for example DNA-squares of a certain size with „nods“ at certain distances. | ||
One member of our team, Daniel Hautzinger, has recently finished his diploma-thesis on Origami-DNA and the possibilities of generating patterns on these square surfaces by modifying the oligo-nucleotides that build up the nod-points.<br> | One member of our team, Daniel Hautzinger, has recently finished his diploma-thesis on Origami-DNA and the possibilities of generating patterns on these square surfaces by modifying the oligo-nucleotides that build up the nod-points.<br> | ||
As the antigens NIP and fluoresceine can as well be fused to these oligos, we had found the seemingly perfect tool to present strictly defined two-dimensional antigen-patterns to cells carrying our synthetic receptor system. | As the antigens NIP and fluoresceine can as well be fused to these oligos, we had found the seemingly perfect tool to present strictly defined two-dimensional antigen-patterns to cells carrying our synthetic receptor system. | ||
| - | + | ||
| - | + | <h2>Methods</h2> | |
| - | + | <h3>Phage DNA<span style="font-weight: bold;"></span></h3> | |
| - | + | <h4><span style="font-weight: bold;"></span><span | |
| - | + | ||
| - | < | + | |
| - | < | + | |
style="font-weight: bold;">Cell culture</span><br> | style="font-weight: bold;">Cell culture</span><br> | ||
| - | </ | + | </h4> |
| - | 50 ml DYT-Medium, | + | 50 ml DYT-Medium, 50 µl tetracycline (TET; 25 mg/ml) and ER2738-cells |
were shaken over night at 37°C. The overnight culture was diluted with | were shaken over night at 37°C. The overnight culture was diluted with | ||
| - | DYT to OD600=0.1 and shaken at 37°C until the culture | + | DYT to OD600=0.1 and shaken at 37°C until the culture reached an OD600 |
around 0.4. Each 50 ml of cell culture were inoculated with 5 µl | around 0.4. Each 50 ml of cell culture were inoculated with 5 µl | ||
M13mp18 phage and shaken for 4 h at 37°C.<br> | M13mp18 phage and shaken for 4 h at 37°C.<br> | ||
| - | < | + | <h4>Isolation of M13mp18 phage from cell culture</h4> |
PEG/NaCl was used to precipitate the phages.<br> | PEG/NaCl was used to precipitate the phages.<br> | ||
<span style="font-weight: bold;">First precipitation</span><br> | <span style="font-weight: bold;">First precipitation</span><br> | ||
| - | Each 50 ml of cell culture | + | Each 50 ml of cell culture was centrifuged at 5000 g for 20 min. While |
| - | the | + | the cells were centrifuged, 1/7 volume PEG/NaCl (about 7ml) was added and |
| - | 7ml | + | transferred in a falcon tube. The phages stay in the |
supernatant therefore the supernatant was carefully decanted to the | supernatant therefore the supernatant was carefully decanted to the | ||
| - | PEG/NaCl and mixed gently by inverting the tube. The mixture ( | + | PEG/NaCl and mixed gently by inverting the tube. The mixture (solution |
1) was left overnight at 4°C. <br> | 1) was left overnight at 4°C. <br> | ||
Solution 1 was centrifuged at 5000 g for 20 min. Because the phage stay | Solution 1 was centrifuged at 5000 g for 20 min. Because the phage stay | ||
in the pellet, the supernatant was removed and the pellet was | in the pellet, the supernatant was removed and the pellet was | ||
| - | resuspended in 2 ml TBS-Buffer ( | + | resuspended in 2 ml TBS-Buffer (solution 2). Solution 2 was put in a |
| - | 1 | + | 1.5 ml Eppendorf tube and centrifuged (13200 rpm, 10 min). After the |
centrifugation the phages stay in the supernatant.<br> | centrifugation the phages stay in the supernatant.<br> | ||
<span style="font-weight: bold;">Second precipitation</span><br> | <span style="font-weight: bold;">Second precipitation</span><br> | ||
170 µl PEG/NaCl(~ 1/6 volume of supernatant) were put in a Eppendorf | 170 µl PEG/NaCl(~ 1/6 volume of supernatant) were put in a Eppendorf | ||
tube. Supernatant was carefully decanted to the PEG/NaCl and mixed | tube. Supernatant was carefully decanted to the PEG/NaCl and mixed | ||
| - | gently by inverting the tube. The mixture ( | + | gently by inverting the tube. The mixture (solution 3) was left for 1 h |
on ice. Solution 3 was centrifuged at 13200 rpm for 10 min.<br> | on ice. Solution 3 was centrifuged at 13200 rpm for 10 min.<br> | ||
| - | < | + | <h4>Measurement of phage titers</h4> |
| - | The absorption of | + | The absorption of solution 3 was measured on a Jasco V-550 UV/VIS |
spectrometer at 269 nm. <br> | spectrometer at 269 nm. <br> | ||
Phage titer was calculated as follows:<br> | Phage titer was calculated as follows:<br> | ||
<span style="font-style: italic;"><span | <span style="font-style: italic;"><span | ||
style="font-weight: bold;">Phage DNA</span> = | style="font-weight: bold;">Phage DNA</span> = | ||
| - | ((A269- | + | ((A269-A320) * 6 * 10^16 * dilution factor) / (number of bases in the |
phage genom = 7249 bp)</span><br> | phage genom = 7249 bp)</span><br> | ||
| - | < | + | <h4>Isolation of the phage DNA </h4> |
The phage DNA was isolated with QIAprep Spin M13-Kit (50) from QIAGEN | The phage DNA was isolated with QIAprep Spin M13-Kit (50) from QIAGEN | ||
(Cat.No: 27704). <br> | (Cat.No: 27704). <br> | ||
DNA-concentration was quantified by Nano-drop photometer.<br> | DNA-concentration was quantified by Nano-drop photometer.<br> | ||
| - | <span style="font-weight: bold;"><br> | + | <span style="font-weight: bold;"><br></span> |
| - | </span> | + | |
| - | < | + | <h3><span style="font-weight: bold;">Origami</span></h3> |
| - | < | + | |
| - | + | <h4><span style="font-weight: bold;"></span>Origami production<br></h4> | |
| - | </ | + | |
To produce the Origami we mixed each the M13mp18 DNA with the oligos, | To produce the Origami we mixed each the M13mp18 DNA with the oligos, | ||
water and TEA/MgAcetat (end concentration =12.5mM).<br> | water and TEA/MgAcetat (end concentration =12.5mM).<br> | ||
[[Image:TeamFreiburg2008_Tabellen-Origamipipettier-Schema2.jpg|800 px]]<br> | [[Image:TeamFreiburg2008_Tabellen-Origamipipettier-Schema2.jpg|800 px]]<br> | ||
Various samples were | Various samples were | ||
| - | produced. For the sample with a ratio of 1:20 (DNA:Oligo) we used 4 nM | + | produced. For the sample with a ratio of 1:20 (DNA:Oligo), we used 4 nM |
| - | DNA and 80 nM of oligos. The Origami were produced in | + | DNA and 80 nM of oligos. The Origami were produced in an eppendorf |
| - | Mastercycler personal. | + | Mastercycler personal. They were heated up to 95°C for 7 min |
and slowly cooled down (0.3°C/s) to 20°C.<br> | and slowly cooled down (0.3°C/s) to 20°C.<br> | ||
Different sample were made:<br> | Different sample were made:<br> | ||
| Line 74: | Line 72: | ||
the 2 oligos with the Alexa 488 were used.<br> | the 2 oligos with the Alexa 488 were used.<br> | ||
<br> | <br> | ||
| - | < | + | <h4><span style="font-weight: bold;"></span>Purification of DNA-Origamis<br> |
| - | </ | + | </h4> |
| - | To purify the DNA-Origamis from the unbound DNA-oligos we used Montage® PCR Centrifugal Filter Devices (Millipore). The Montage® PCR Centrifugal Filter Devices were labeled and put with the purple side on | + | To purify the DNA-Origamis from the unbound DNA-oligos, we used Montage® PCR Centrifugal Filter Devices (Millipore). The Montage® PCR Centrifugal Filter Devices were labeled and put with the purple side on top in 1.5 ml Eppendorf tubes. To clean the filter of remaining glycerol, 450 µl TAE/MgAcetat (12.5 mM; 1x filtered) was put on top of the filter and centrifuged for 15 min at 1000 g. After removing the filtrate, 400 µl TEA/MgAcetat (12.5 mM;1x filtered) and 45 µl DNA-origami were put on top of the filter and again centrifuged for 15 min at 1000 g. The filtrate was removed again. All unbound DNA-oligos were washed off by putting 400 µl TEA/MgAcetat (12.5 mM; 1x filtered) on top of the filter. The sample was centrifuged for 15 min at 1000 g. To release the DNA-origamis of the filter, 100 µl TAE/MgAcetat (12.5 mM;1x filtered) was put on top of the filter, and the filter was left at room temperature for at least 2 min. The filter shouldn´t run dry. The Montage® PCR Centrifugal Filter Devices were put upside down (the purple side has to be on the bottom) in one of the special Invert Spin tubes form Millipore and centrifuged for 3 min at 1000 g. |
| - | The Origami were kept in different buffers. For this TEA/MgAcetat (12.5 mM; 1x filtered) was replaced by the according buffer.<br> | + | The Origami were kept in different buffers. For this, TEA/MgAcetat (12.5 mM; 1x filtered) was replaced by the according buffer.<br> |
| - | < | + | <h4><span style="font-weight: bold;"></span>Atomic force microscopy to test the origami stability |
<br> | <br> | ||
| - | </ | + | </h4> |
To see if the Origami were formed well and stable in the different buffers an atomic force microscope (AFM) was used. The measurement itself was done in air (not in the buffer). | To see if the Origami were formed well and stable in the different buffers an atomic force microscope (AFM) was used. The measurement itself was done in air (not in the buffer). | ||
| - | The DNA-Origami were absorbed to freshly cleaved mica. Therefore the mica was cut into 6 mm pieces and affixed to the metal panes we used for the measurement. To get | + | The DNA-Origami were absorbed to freshly cleaved mica. Therefore the mica was cut into 6 mm pieces and affixed to the metal panes we used for the measurement. To get an atomically clean surface, an adhesive tape was used to remove the topmost mica layers. After this the sample could be put on. First 2-10 µl of the sample were put on the mica and then quickly diluted with water (just as much that the mica was covered with fluid). The sample was incubated for about 5 min and then the mica was blown dry with a stream of nitrogen. Then the sample could be measured. |
The metal pane was fixed in the metal sample holder by a magnet, so that the sample could not move itself during the measurement. | The metal pane was fixed in the metal sample holder by a magnet, so that the sample could not move itself during the measurement. | ||
| - | < | + | <h2>Results and Discussion</h2> |
| - | < | + | <h3>1. Different ratios of phage DNA to staple oligonucleotides</h3> |
Because we also wanted to measure the calcium influx in the LSRII | Because we also wanted to measure the calcium influx in the LSRII | ||
fluorescence spectrometer, we had to increase the concentration of the | fluorescence spectrometer, we had to increase the concentration of the | ||
Origamis at least up to 200 nM. Because the oligos we had to use for | Origamis at least up to 200 nM. Because the oligos we had to use for | ||
| - | building | + | building DNA-Origami are very expensive, we first tried to reduce the |
| - | ratio of | + | ratio of phage DNA to oligos. Therefore we tried to make Origamis with two |
(1:10) and four (1:5) times lower concentration of oligos. As positive | (1:10) and four (1:5) times lower concentration of oligos. As positive | ||
| - | control we took the 1:20 ratio at which we | + | control we took the 1:20 ratio at which we knew it should be stable. We |
| - | used | + | used AFM to check if the Origami are well formed. The results are |
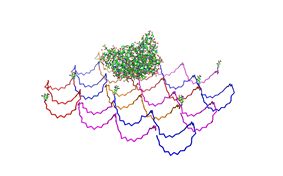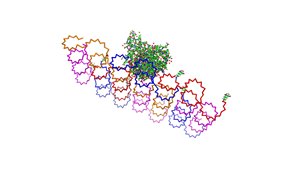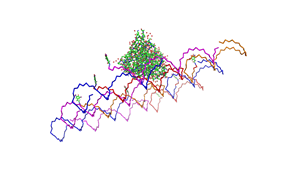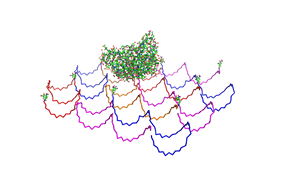
shown in figures 1-3. <br> | shown in figures 1-3. <br> | ||
<br> | <br> | ||
| - | [[Image: | + | [[Image:TeamFreiburg2008-1zu5_10_20.png|700 px]] |
| - | + | ||
| - | + | ||
<br> | <br> | ||
| - | As we see in the | + | <br> |
| + | As we see in the Figures 1-3, all Origami are well formed. So we are | ||
able to use a 1:5 ratio to produce our Origami.<br> | able to use a 1:5 ratio to produce our Origami.<br> | ||
| - | < | + | <h3>2. Origami in Krebs-Ringer-Hepes buffer</h3> |
The TEA/MgAcetat buffer we used to build and keep the Origami does not | The TEA/MgAcetat buffer we used to build and keep the Origami does not | ||
have any salts beside the magnesium. Therefore the T cells are not able | have any salts beside the magnesium. Therefore the T cells are not able | ||
to survive long enough to measure the calcium influx.<br> | to survive long enough to measure the calcium influx.<br> | ||
Beside that, the EDTA would disturb the calcium measurement. Hence we | Beside that, the EDTA would disturb the calcium measurement. Hence we | ||
| - | + | had to find a different buffer in which the Origami and the T | |
cells are stable and which can also be used for the calcium | cells are stable and which can also be used for the calcium | ||
measurement. In literature we found, that many people use a so-called | measurement. In literature we found, that many people use a so-called | ||
Krebs-Ringer-HEPES buffer for calcium measurement in LSRII fluorescence | Krebs-Ringer-HEPES buffer for calcium measurement in LSRII fluorescence | ||
| - | spectrometer. We | + | spectrometer. We used 1:20 and 1:5 ratio of DNA to oligo to |
| - | make the | + | make the Origami. Each of the ratios was buffered in Krebs-ringer-HEPES |
buffer. As positive control we used Origami buffered in TAE/MgAc. The | buffer. As positive control we used Origami buffered in TAE/MgAc. The | ||
results are shown in figure 4 and 5.<br> | results are shown in figure 4 and 5.<br> | ||
<br> | <br> | ||
| - | + | [[Image:Team_Freiburg2008_Bilder-vom-07-23-2008_1zu20_kontrolle_und_Ringer.jpg|800 px]]<br> | |
| + | <small>Fig. 4:DNA-Origami(1:20) in TAE/MgAcetat (control) in Krebs-Ringer-Hepes buffer</small><br> | ||
| + | <br> | ||
| + | [[Image:Team_Freiburg2008_Bilder-vom-07-23-2008_1zu5_kontrolle_und_Ringer.jpg|800 px]] | ||
| + | <small>Fig. 5:DNA-Origami(1:5) in TAE/MgAcetat (control) in Krebs-Ringer-Hepes buffer</small><br> | ||
<br> | <br> | ||
Both samples in Krebs-Ringer-HEPES buffer did not show the completely | Both samples in Krebs-Ringer-HEPES buffer did not show the completely | ||
formed Origami. Some bigger structures in Krebs-Ringer-HEPES buffer in | formed Origami. Some bigger structures in Krebs-Ringer-HEPES buffer in | ||
figure 4 seem to be parts of the Origami. Maybe the Origami did form | figure 4 seem to be parts of the Origami. Maybe the Origami did form | ||
| - | right in the MasterCycler, but then | + | right in the MasterCycler, but then fell apart when we buffered them in |
the Krebs-Ringer-HEPES buffer. Because we read in literature that some | the Krebs-Ringer-HEPES buffer. Because we read in literature that some | ||
salts in the Krebs-Ringer-HEPES buffer could disturb the interaction of | salts in the Krebs-Ringer-HEPES buffer could disturb the interaction of | ||
| Line 129: | Line 130: | ||
factor in the Krebs-Ringer-HEPES buffer could be the lack of magnesium | factor in the Krebs-Ringer-HEPES buffer could be the lack of magnesium | ||
or the calcium. Therefore we first tested if the Origami are stable in | or the calcium. Therefore we first tested if the Origami are stable in | ||
| - | Origami in Krebs-Ringer-Hepes buffer with 12. | + | Origami in Krebs-Ringer-Hepes buffer with 12.5 mM magnesium. Still we |
didn´t see any Origami (data not shown). <br> | didn´t see any Origami (data not shown). <br> | ||
<br> | <br> | ||
| - | < | + | <h3>3. Future prospects</h3> |
Calcium has the same charge as magnesium, but the ionic radius of | Calcium has the same charge as magnesium, but the ionic radius of | ||
magnesium is much bigger, which could lead to deformed and instable | magnesium is much bigger, which could lead to deformed and instable | ||
Origami. Therefore it could be also tested if the Origami are stable in | Origami. Therefore it could be also tested if the Origami are stable in | ||
| - | Krebs-Ringer-HEPES buffer with 12. | + | Krebs-Ringer-HEPES buffer with 12.5 mM magnesium, but without calcium. |
Because the buffer has also to be suitable for the cells, the stability | Because the buffer has also to be suitable for the cells, the stability | ||
of the Origami in phosphate buffer without calcium should be tested.<br> | of the Origami in phosphate buffer without calcium should be tested.<br> | ||
<br> | <br> | ||
| - | + | <h2>[[Image:MO2.jpg|50px|]]Literature</h2> | |
| + | *Paul W. K. Rothemund:"Folding DNA to create nanoscale shapes and patterns", Nature 440, 297-302 (16 March 2006) | ||
}} | }} | ||
Latest revision as of 01:50, 30 October 2008
 "
"