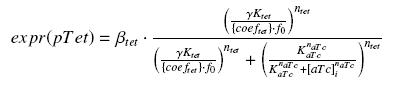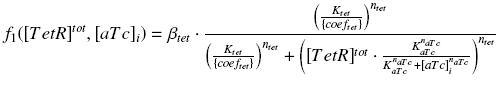|
|
| (8 intermediate revisions not shown) |
| Line 2: |
Line 2: |
| | | | |
| | {{Paris/Header|Method & Algorithm : ƒ1}} | | {{Paris/Header|Method & Algorithm : ƒ1}} |
| | + | <center> = act_''pTet'' </center> |
| | + | <br> |
| | | | |
| | [[Image:f1a.jpg|thumb|Specific Plasmid Characterisation for ƒ1]] | | [[Image:f1a.jpg|thumb|Specific Plasmid Characterisation for ƒ1]] |
| | | | |
| - | The experience would give us
| + | According to the characterization plasmid (see right) and to our modeling, in the '''exponential phase of growth''', at the steady state, the experiment would give us |
| | | | |
| | [[Image:f1expr.jpg|center]] | | [[Image:f1expr.jpg|center]] |
| | | | |
| - | Thus, at steady-state and in the exponential phase of growth :
| + | and at steady-state and in the exponential phase of growth, we expect : |
| | | | |
| | [[Image:ExprptetF0.jpg|center]] | | [[Image:ExprptetF0.jpg|center]] |
| | | | |
| | + | we use this analytical expression to determine the parameters : |
| | | | |
| | <div style="text-align: center"> | | <div style="text-align: center"> |
| - | {{Paris/Toggle|Table|Team:Paris/Modeling/More_f1_Table}} | + | {{Paris/Toggle|Table of Values|Team:Paris/Modeling/More_f1_Table}} |
| | </div> | | </div> |
| - |
| |
| | | | |
| | <div style="text-align: center"> | | <div style="text-align: center"> |
| - | {{Paris/Toggle|Algorithm|Team:Paris/Modeling/More_f1_Algo}} | + | {{Paris/Toggle|Algorithms|Team:Paris/Modeling/More_f1_Algo}} |
| | </div> | | </div> |
| | | | |
| - | {|border="1" style="text-align: center"
| + | Also, this experiment will enable us to know the expression of ƒ1 : |
| - | |param
| + | |
| - | |signification
| + | |
| - | |unit
| + | |
| - | |value
| + | |
| - | |comments
| + | |
| - | |-
| + | |
| - | |[expr(pTet)]
| + | |
| - | |expression rate of <br> pTet '''with RBS E0032'''
| + | |
| - | |nM.min<sup>-1</sup>
| + | |
| - | |
| + | |
| - | |need for 20 values with well choosen [aTc]<sub>i</sub> <br>
| + | |
| - | with 5 measures for each
| + | |
| - | |-
| + | |
| - | |γ<sub>GFP</sub>
| + | |
| - | |dilution-degradation rate <br> of GFP(mut3b)
| + | |
| - | |min<sup>-1</sup>
| + | |
| - | |0.0198
| + | |
| - | |Time for Cell Division : 35 min. ; No degradation
| + | |
| - | |-
| + | |
| - | |[GFP]
| + | |
| - | |GFP concentration at steady-state
| + | |
| - | |nM
| + | |
| - | |
| + | |
| - | |need for 100 measures
| + | |
| - | |-
| + | |
| - | |(''fluorescence'')
| + | |
| - | |value of the observed fluorescence
| + | |
| - | |au
| + | |
| - | |
| + | |
| - | |need for 100 measures
| + | |
| - | |-
| + | |
| - | |''conversion''
| + | |
| - | |conversion ration between <br> fluorescence and concentration
| + | |
| - | |nM.au<sup>-1</sup>
| + | |
| - | |(1/79.429)
| + | |
| - | |
| + | |
| - | |}
| + | |
| | | | |
| - | <br><br>
| + | [[Image:ExprF1.jpg|center]] |
| | | | |
| - | {|border="1" style="text-align: center"
| + | <br> |
| - | |param
| + | |
| - | |signification <br> corresponding parameters in the [[Team:Paris/Modeling/Oscillations#Resulting_Equations|equations]]
| + | |
| - | |unit
| + | |
| - | |value
| + | |
| - | |comments
| + | |
| - | |-
| + | |
| - | |β<sub>tet</sub>
| + | |
| - | |production rate of pTet '''with RBS E0032''' <br> β<sub>1</sub>
| + | |
| - | |nM.min<sup>-1</sup>
| + | |
| - | |
| + | |
| - | |
| + | |
| - | |-
| + | |
| - | |(K<sub>tet</sub>/{coef<sub>tet</sub>})
| + | |
| - | |activation constant of pTet <br> K<sub>20</sub>
| + | |
| - | |nM
| + | |
| - | |
| + | |
| - | |The [[Team:Paris/Modeling/Codes|optimisation program]] will give us (γ K<sub>tet</sub> / {coef<sub>tet</sub>} ƒ0) <br> The literature [[Team:Paris/Modeling/Bibliography|[?] ]] gives K<sub>tet</sub> =
| + | |
| - | |-
| + | |
| - | |n<sub>tet</sub>
| + | |
| - | |complexation order of pTet<br> n<sub>20</sub>
| + | |
| - | |no dimension
| + | |
| - | |
| + | |
| - | |The literature [[Team:Paris/Modeling/Bibliography|[?] ]] gives n<sub>tet</sub> =
| + | |
| - | |-
| + | |
| - | |K<sub>aTc</sub>
| + | |
| - | |complexation constant aTc-TetR <br> K<sub>19</sub>
| + | |
| - | |nM
| + | |
| - | |
| + | |
| - | |The literature [[Team:Paris/Modeling/Bibliography|[?] ]] gives K<sub>aTc</sub> =
| + | |
| - | |-
| + | |
| - | |n<sub>aTc</sub>
| + | |
| - | |complexation order aTc-TetR <br> n<sub>19</sub>
| + | |
| - | |no dimension
| + | |
| - | |
| + | |
| - | |The literature [[Team:Paris/Modeling/Bibliography|[?] ]] gives n<sub>aTc</sub> =
| + | |
| - | |}
| + | |
| | | | |
| - | Also, this experiment will allow us to know the expression of ƒ1 :
| + | <center> |
| - | | + | [[Team:Paris/Modeling/Implementation| <Back - to "Implementation" ]]| <br> |
| - | [[Image:ExprF1.jpg|center]] | + | [[Team:Paris/Modeling/Protocol_Of_Characterization| <Back - to "Protocol Of Characterization" ]]| |
| | + | </center> |
|
Method & Algorithm : ƒ1
= act_pTet
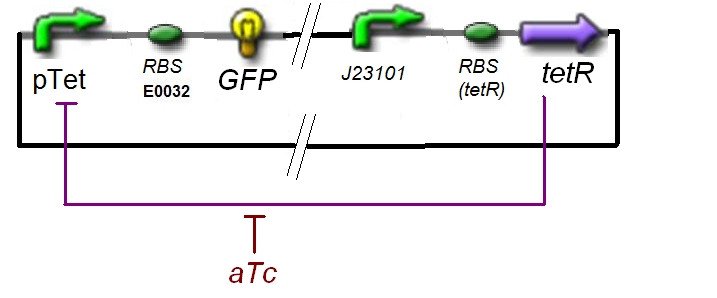 Specific Plasmid Characterisation for ƒ1 According to the characterization plasmid (see right) and to our modeling, in the exponential phase of growth, at the steady state, the experiment would give us
and at steady-state and in the exponential phase of growth, we expect :
we use this analytical expression to determine the parameters :
↓ Table of Values ↑
| param
| signification
| unit
| value
| comments
|
| (fluorescence)
| value of the observed fluorescence
| au
|
| need for 20 values with well choosen [aTc]i
|
| conversion
| conversion ration between
fluorescence and concentration
↓ gives ↓
| nM.au-1
| (1/79.429)
|
|
| [GFP]
| GFP concentration at steady-state
| nM
|
|
|
| γGFP
| dilution-degradation rate
of GFP(mut3b)
↓ gives ↓
| min-1
| 0.0198
| Only dilution :
Time Cell Division : 35 min.
|
| ƒ1
| activity of
pTet with RBS E0032
| nM.min-1
|
|
|
| param
| signification
corresponding parameters in the equations
| unit
| value
| comments
|
| βtet
| basal activity of
pTet with RBS E0032
β16
| nM.min-1
|
|
|
| (Ktet/{coeftetR})
| activation constant of TetR><pTet
K13
| nM
|
| The optimisation program will give us (γ Ktet / {coeftet} ƒ0)
The literature [?] gives Ktet =
|
| ntet
| complexation order of TetR><pTet
n13
| no dimension
|
| The literature [?] gives ntet =
|
| KaTc
| complexation constant aTc><TetR
K12
| nM
|
| The literature [?] gives KaTc =
|
| naTc
| complexation order aTc><TetR
n12
| no dimension
|
| The literature [?] gives naTc =
|
|
↓ Algorithms ↑
|
find_ƒ1
function optimal_parameters = find_f1(X_data, Y_data, initial_parameters)
function output = expr_pTet(parameters, X_data)
for k = 1:length(X_data)
output(k) = parameters(1) * (1 - ...
hill((1 - hill(X_data(k),parameters(4),parameters(5))),parameters(2),parameters(3)));
end
end
options=optimset('LevenbergMarquardt','on','TolX',1e-10,'MaxFunEvals',1e10,'TolFun',1e-10,'MaxIter',1e4);
optimal_parameters = lsqcurvefit( @(parameters, X_data) expr_pTet(parameters, X_data), ...
initial_parameters, X_data, Y_data, 1/10*initial_parameters, 10*initial_parameters, options );
end
Inv_ƒ1
function quant_aTc = Inv_f1(inducer_quantity)
global gamma, f0;
function equa = F(x)
equa = f1( (f0/gamma) , x ) - inducer_quantity;
end
options=optimset('LevenbergMarquardt','on','TolX',1e-10,'MaxFunEvals',1e10,'TolFun',1e-10,'MaxIter',1e4);
quant_aTc = fsolve(F,1,options);
end
|
Also, this experiment will enable us to know the expression of ƒ1 :
<Back - to "Implementation" |
<Back - to "Protocol Of Characterization" |
|
 "
"