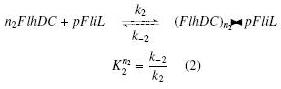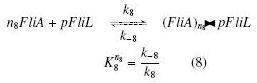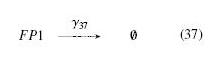Team:Paris/Modeling/Molecular Reactions
From 2008.igem.org
(Difference between revisions)
| (49 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Paris/Menu}} | {{Paris/Menu}} | ||
| + | |||
{{Paris/Header|Molecular Reactions}} | {{Paris/Header|Molecular Reactions}} | ||
| + | {{Paris/Section_contents_characterization}} | ||
| + | |||
| + | We show here the elementary reactions that served us as basis. They intend to be "exhaustive" in the sense that every step of the [[Team:Paris/Modeling/Protocol_Of_Characterization|system]] is described. Each of these reactions has to be "mathematically interpreted", in a [[Team:Paris/Modeling/FromMolReactToNLOde|model]] which accounts for the observable phenomena, and, in the same time, allows us to well estimate [[Team:Paris/Modeling/Protocol_Of_Characterisation|experimentally]] all the involved parameters. | ||
<br> | <br> | ||
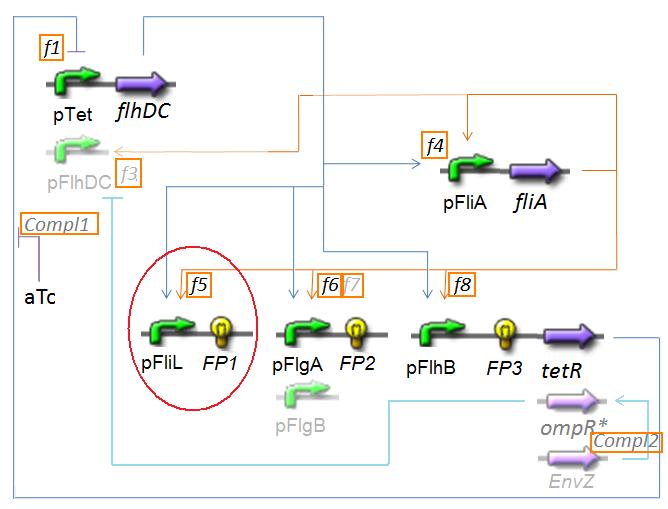
| - | + | '''The system we are studying is the "Core System with three genes of interest"''' that we can represent in the scheme below (with all alternatives ''pFlhDC'' or ''pTet'' and ''TetR'' or ''envZ'' or ''OmpR<sup>*</sup>'') | |
| + | |||
| + | <div style="text-align: center"> | ||
| + | {{Paris/Toggle|Core System|Team:Paris/Modeling/More_Mol_Reac_System|200px}} | ||
| + | </div> | ||
<br> | <br> | ||
| - | ''' | + | {| border="1" width="100%" style="text-align: center" |
| + | |- | ||
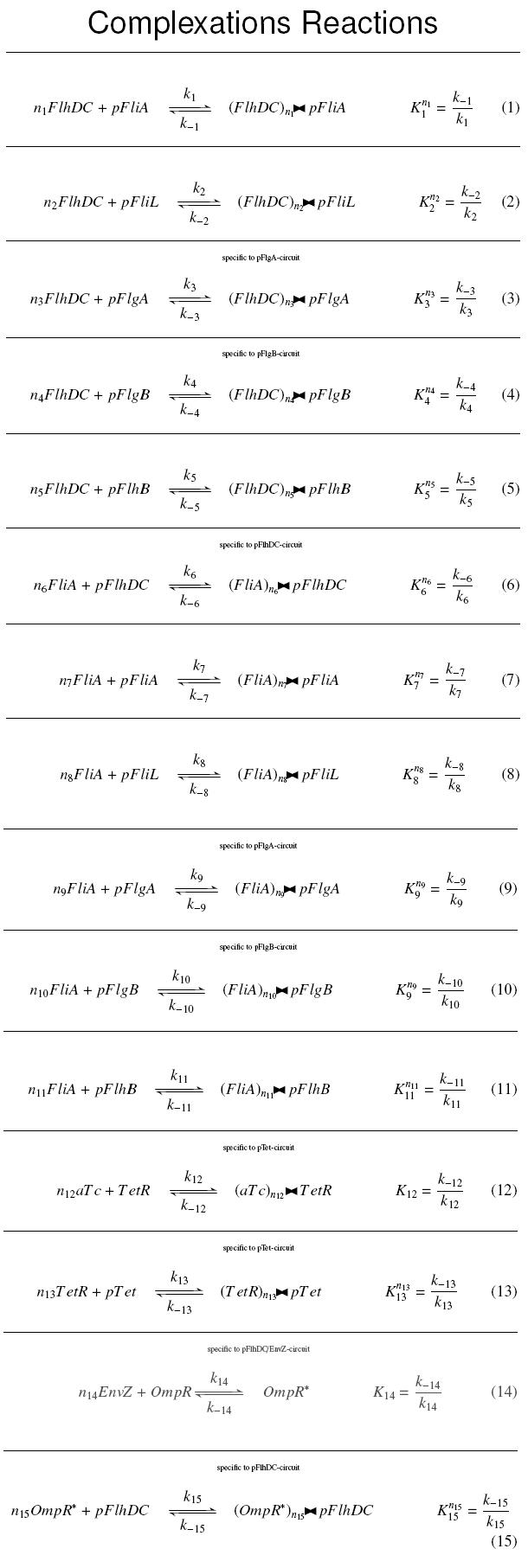
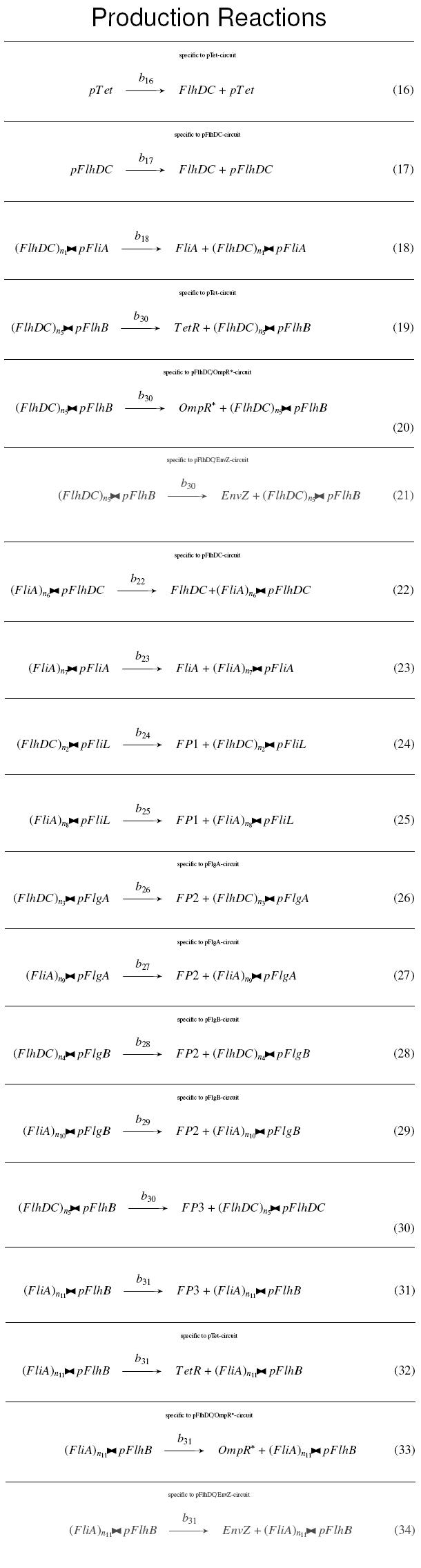
| + | | width="75%" | '''Complexation Reactions''' <br> | ||
| + | The double triangle ('''><''') is a symbol for complexations : <br> | ||
| + | [[Image:compl.jpg|110px]] means ''n<sub>1</sub>'' molecules of ''FlhDC'' complexed with a ''pFliA'' promoter<br><br> | ||
| - | <br><br> | + | <div style="text-align: center"> |
| + | {{Paris/Toggle2|Complexation Reactions|Team:Paris/Modeling/Compl_React}} | ||
| + | </div> | ||
| + | |||
| + | | width="25%" | '''On the Example :''' <br> | ||
| + | <small> How given amounts of ''FlhDC'' and ''FliA'' would produce ''FP1'' ? </small><br> | ||
| + | <br> | ||
| + | * ''FlhDC'' and ''FliA'' bind to ''pFliL'' : <br> | ||
| + | [[Image:DCiLcompl.jpg]] | ||
| + | |||
| + | [[Image:AiLcompl.jpg]] | ||
| - | |||
|- | |- | ||
| - | |Reactions | + | | width="75%" | '''Production Reactions''' <br> |
| - | |On the Example [[Image: | + | The double triangle ('''><''') is a symbol for complexations : <br> |
| + | [[Image:compl.jpg|110px]] means ''n<sub>1</sub>'' molecules of ''FlhDC'' complexed with a ''pFliA'' promoter<br><br> | ||
| + | |||
| + | <div style="text-align: center"> | ||
| + | {{Paris/Toggle2|Production Reactions|Team:Paris/Modeling/Prod_React}} | ||
| + | </div> | ||
| + | |||
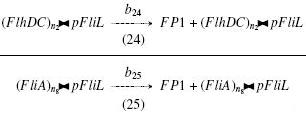
| + | | width="25%" | '''On the Example :''' <br> | ||
| + | <small> How given amounts of ''FlhDC'' and ''FliA'' would produce ''FP1'' ? </small> <br> | ||
| + | <br> | ||
| + | * ''FlhDC><pFliL'' and ''FliA><pFliL'' causes "production" of ''FP1'' : <br> | ||
| + | <br> <br> | ||
| + | [[Image:bFP1.jpg]] | ||
| + | |||
|- | |- | ||
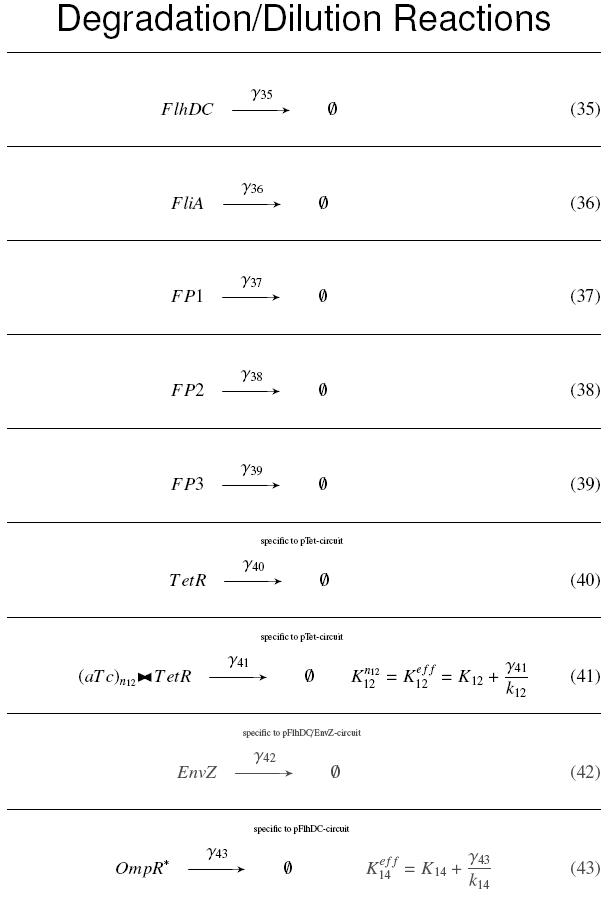
| - | | | + | | width="75%" | '''Degradation/Dilution Reactions''' <br><br> |
| - | | | + | |
| - | + | <div style="text-align: center"> | |
| - | | | + | {{Paris/Toggle2|Degradation/Dilution Reactions|Team:Paris/Modeling/D_React}} |
| - | | | + | </div> |
| - | + | ||
| - | + | | width="25%" | '''On the Example :''' <br> | |
| - | | | + | <small> How given amounts of ''FlhDC'' and ''FliA'' would produce ''FP1'' ? </small><br> |
| + | <br> | ||
| + | * ''FP1'' disappears because of its degradation and dilution : <br> | ||
| + | [[Image:dFP1.jpg]] | ||
| + | |||
|} | |} | ||
| + | <div style="text-align: right">For complete PDF with all Reactions and Numbered References : [https://static.igem.org/mediawiki/2008/6/61/Equa_Core_System.pdf complete list of reactions and equations] </div> | ||
| + | |||
| + | <br> | ||
| + | |||
| + | {{Paris/Navig|Team:Paris/Modeling/Workflow_Example}} | ||
Latest revision as of 03:39, 30 October 2008
|
Molecular Reactions
Other pages:
We show here the elementary reactions that served us as basis. They intend to be "exhaustive" in the sense that every step of the system is described. Each of these reactions has to be "mathematically interpreted", in a model which accounts for the observable phenomena, and, in the same time, allows us to well estimate experimentally all the involved parameters.
The system we are studying is the "Core System with three genes of interest" that we can represent in the scheme below (with all alternatives pFlhDC or pTet and TetR or envZ or OmpR*) ↓ Core System ↑
For complete PDF with all Reactions and Numbered References : complete list of reactions and equations
|
 "
"