Team:BrownTwo/Implementation/construction
From 2008.igem.org
(→Yeast Integration) |
m (→Fluorescent Tagging of Transcription Factors) |
||
| (One intermediate revision not shown) | |||
| Line 21: | Line 21: | ||
==Fluorescent Tagging of Transcription Factors== | ==Fluorescent Tagging of Transcription Factors== | ||
| - | + | We received our library of transcription factors as fusion proteins tagged with the mCherry fluorescent domain. As we wish to keep track of the concentration of a number of different elements in our network, we elected to construct repressors bearing the cyan fluorescent protein CFP, which can be distinguished from the mCherry fluorescence from the other repressors and the YFP fluorescent reporter that we use as an output. We removed the fluorescence from our activator, as we can infer its concentration from the level of input. With CFP on the tau repressor and mCherry on alpha and R (both under the same promoter), we hope to get a solid understanding of the dynamics of each element of our system given varying levels of induction and different thresholds. | |
==Yeast Integration== | ==Yeast Integration== | ||
| - | + | In order to easily test many variations on the limiter circuit, we ligated variations on each element onto the same pRS30x series integrative vector. Thus, we can create a strain bearing the A, alpha, R, and YFP reporter, and later ligate in a variety of threshold-setting tau constructs to test their effects. As of press time, we have generated the library of yeast constructs and are ready to transform our limiter. We hope to get some results in time for the Jamboree. | |
Latest revision as of 04:06, 30 October 2008
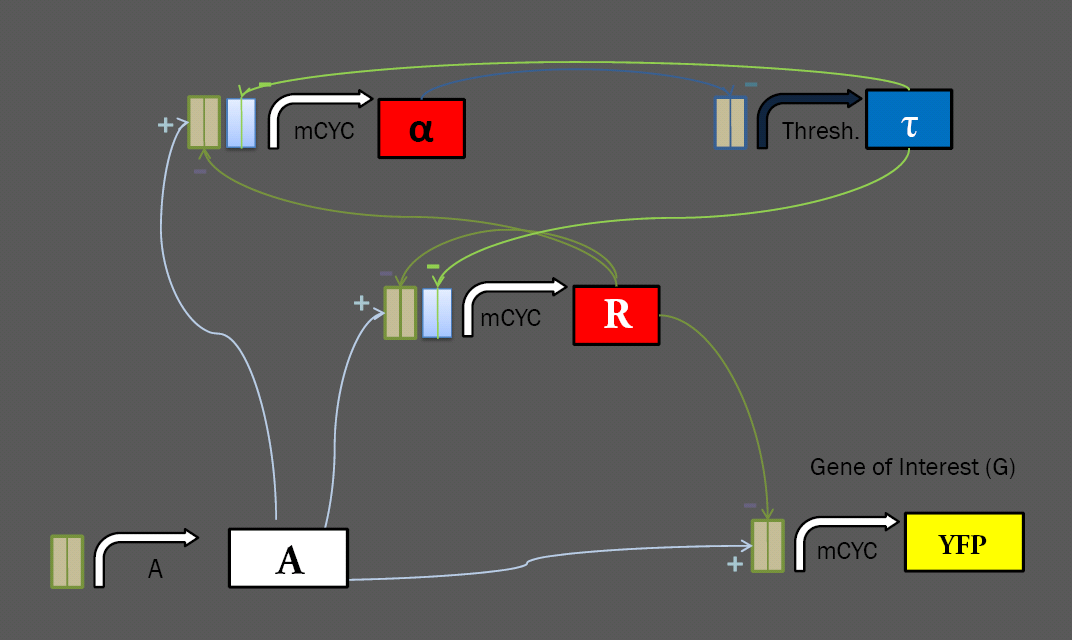
A Modular Approach to Constructing the Circuit
The general layout of our limiter circuit is very amenable to variation. Each of the logical roles can be filled by a number of different parts, possibly resulting in different behavior. This process was made especially straightforward by the modular transcriptional control elements at our disposal. Work with these transcription factors in the Silver Lab at Harvard showed that they show efficacy in regulating a variety of promoters, including the minimal mCYC1 and the inducible pGAL1. To extend this work further, we have constructed a library of regulable promoters for use in our network, ligating binding sites for the synthetic transcription factors onto a variety of yeast promoters. Regulatory RegionsMinimal PromotersmCYC was used as a minimal promoter, with all transcriptional induction and repression coming from transcription factors binding to sites ligated upstream. We generated a variety of minimal promoters for use in the limiter system, with mCYC ligated to each of the four sets of binding sites, and three variations on mCYC ligated to two different sets of binding sites. These will allow for construction of multiple variations on the limiter circuit. Constitutive PromotersIn a limiter circuit with a static threshold, the Thresh. promoter above is constitutive, but also requires regulation from alpha. To investigate the effect of changing the threshold, we ligated binding sites onto two constitutive promoters: the high-level TEF2, and the low-level ADH1. Repressable PromotersWork in the Silver Lab has shown success in using binding sites to regulate inducible promoters. We have constructed two versions of the MET25 repressable promoter with binding sites, hoping to construct a limiter circuit in which we can set the threshold by varying the input concentration of methionine. Fluorescent Tagging of Transcription FactorsWe received our library of transcription factors as fusion proteins tagged with the mCherry fluorescent domain. As we wish to keep track of the concentration of a number of different elements in our network, we elected to construct repressors bearing the cyan fluorescent protein CFP, which can be distinguished from the mCherry fluorescence from the other repressors and the YFP fluorescent reporter that we use as an output. We removed the fluorescence from our activator, as we can infer its concentration from the level of input. With CFP on the tau repressor and mCherry on alpha and R (both under the same promoter), we hope to get a solid understanding of the dynamics of each element of our system given varying levels of induction and different thresholds. Yeast IntegrationIn order to easily test many variations on the limiter circuit, we ligated variations on each element onto the same pRS30x series integrative vector. Thus, we can create a strain bearing the A, alpha, R, and YFP reporter, and later ligate in a variety of threshold-setting tau constructs to test their effects. As of press time, we have generated the library of yeast constructs and are ready to transform our limiter. We hope to get some results in time for the Jamboree. |
 "
"