Team:Harvard/Project
From 2008.igem.org
(→Lac-inducible Strains) |
m |
||
| (One intermediate revision not shown) | |||
| Line 5: | Line 5: | ||
table { | table { | ||
background-color: #c4dbea; | background-color: #c4dbea; | ||
| - | font-color: | + | font-color: #cccccc; |
color:white; | color:white; | ||
} | } | ||
| Line 14: | Line 14: | ||
} | } | ||
| + | .firstHeading { | ||
| + | color:white; | ||
} | } | ||
| + | |||
| + | #bodyContent { | ||
| + | background-color: #c4dbea; | ||
| + | } | ||
#content { | #content { | ||
| Line 47: | Line 53: | ||
<!--- body here---> | <!--- body here---> | ||
| - | {|align="justify" style="background-color:#FFFFFF;text-indent: 15pt;text-align:justify" cellpadding="50" width="90%" | + | {|align="justify" style="background-color:#FFFFFF;text-indent: 15pt; font-color:#cccccc; text-align:justify" cellpadding="50" width="90%" |
| | | | ||
==BACTRICITY: Bacterial Biosensors with Electrical Output== | ==BACTRICITY: Bacterial Biosensors with Electrical Output== | ||
| Line 127: | Line 133: | ||
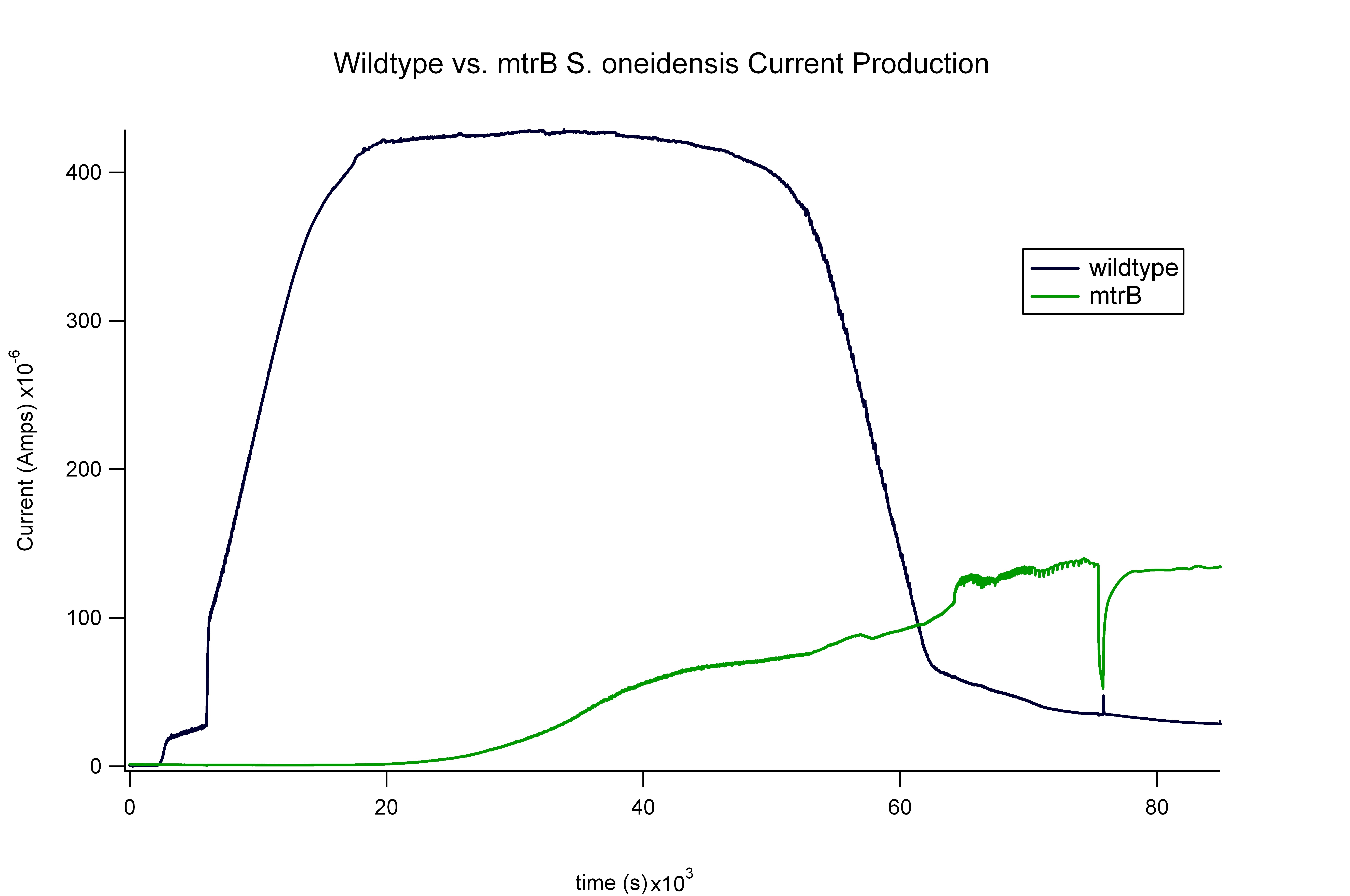
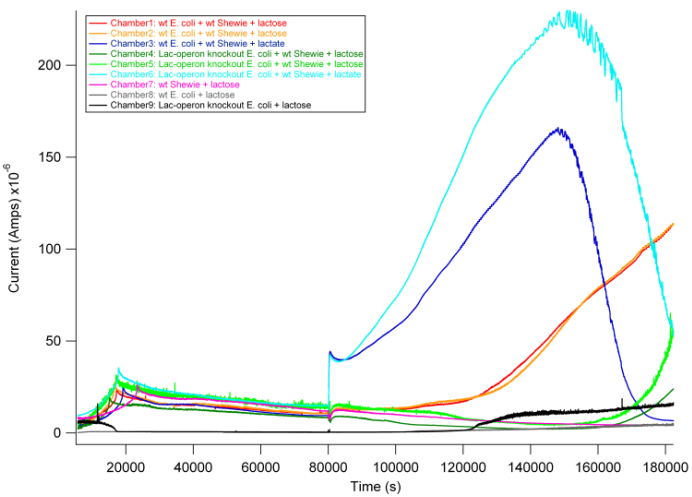
Based on previous experiments, we would expect current production in combinations 2 and 4 as they both have wildtype S. oneidensis MR-1 and receive lactate. In addition, however, we would expect combination 1 to also produce current. As described above, wt E. coli would break down lactose into lactate, and S. oneidensis MR-1 would break down lactate to produce current. | Based on previous experiments, we would expect current production in combinations 2 and 4 as they both have wildtype S. oneidensis MR-1 and receive lactate. In addition, however, we would expect combination 1 to also produce current. As described above, wt E. coli would break down lactose into lactate, and S. oneidensis MR-1 would break down lactate to produce current. | ||
| - | [[ Image: picture 5.png | | + | [[ Image: picture 5.png | 750px ]] |
From the data above, we found that combination 1 did indeed produce current with a delay relative to the positive control. The delay can be attributed to the time it takes for E. coli to break down lactose into lactate, thus adding an extra step in the carbon source to current production pathway compared to our positive controls. These results are exciting in that they show a possibility of taking advantage of this cooperative effort to achieve inducible current. | From the data above, we found that combination 1 did indeed produce current with a delay relative to the positive control. The delay can be attributed to the time it takes for E. coli to break down lactose into lactate, thus adding an extra step in the carbon source to current production pathway compared to our positive controls. These results are exciting in that they show a possibility of taking advantage of this cooperative effort to achieve inducible current. | ||
| Line 135: | Line 141: | ||
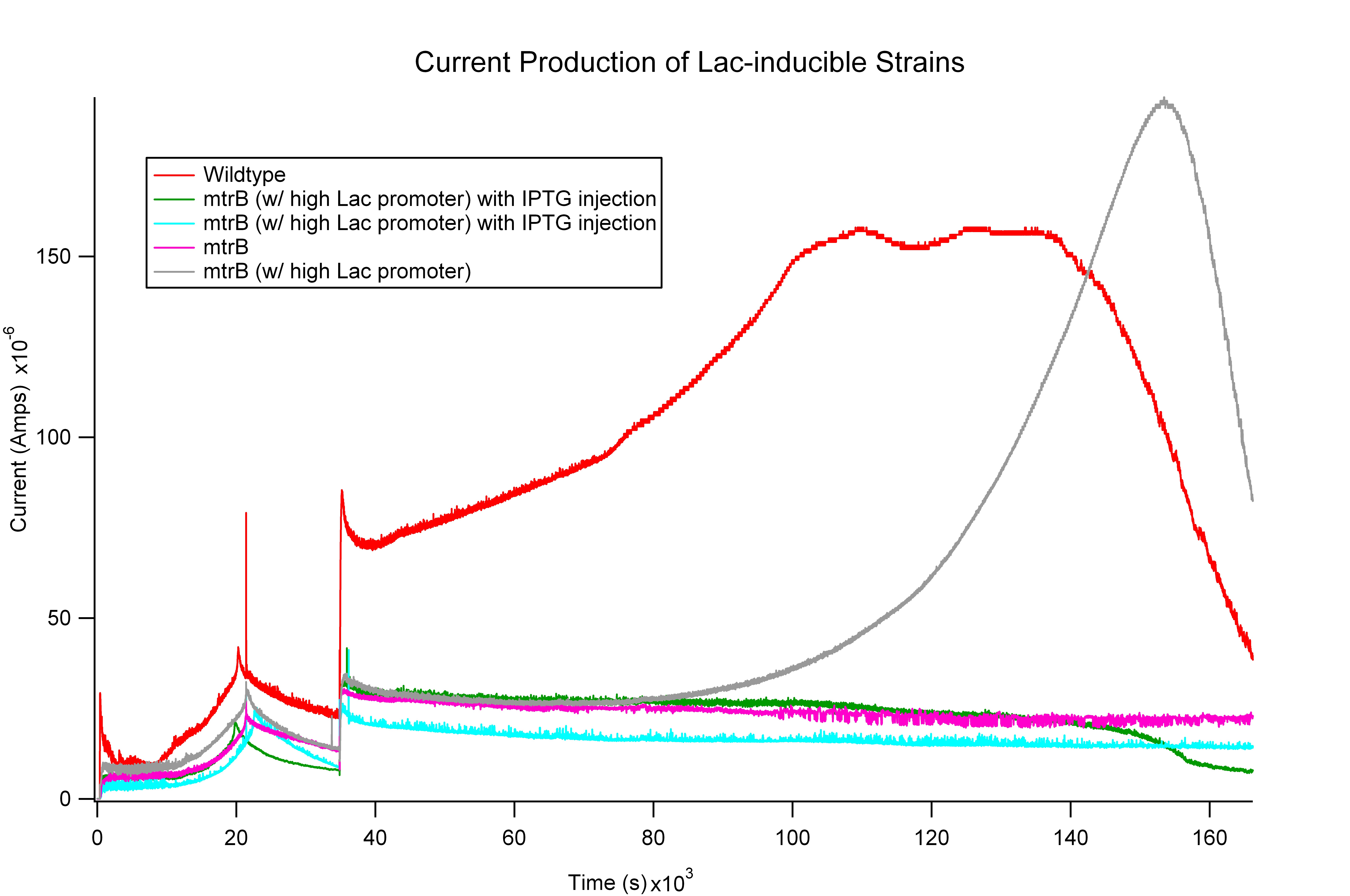
Our work with creating a system of inducible electrical output in ''S. oneidensis'' has laid the foundations for many different exciting avenues of further inquiry which look to take advantage of a bacteria-computer interface that combines the amazing sensitivity and adaptability of bacteria with the speed and analytical abilities of electricity and computers. | Our work with creating a system of inducible electrical output in ''S. oneidensis'' has laid the foundations for many different exciting avenues of further inquiry which look to take advantage of a bacteria-computer interface that combines the amazing sensitivity and adaptability of bacteria with the speed and analytical abilities of electricity and computers. | ||
| - | Using the same principles underlying the lac system, the [ | + | Using the same principles underlying the lac system, the [https://2006.igem.org/University_of_Edinburgh_2006 arsenic biosensor] developed by the University of Edinburgh iGEM 2006 team could be introduced into ''S. oneidensis'', allowing for the coupling of arsenic sensing to an electrical output, a form of a data which is easier to automate and transmit. This could be further extended to other chemical sensing systems, such as the [https://2007.igem.org/Brown lead sensor] created by the Brown iGEM 2007 team and the [https://2007.igem.org/MIT mercury sensor] made by the MIT iGEM 2007 team, resulting ultimately in an array of different strains of ''S. oneidensis'' which all respond to the presence of different chemicals with an electrical output that can be monitored by a computer. This could theoretically allow for the remote sensing and analysis of the chemical composition of an environment over time in a cost-effective manner, making it a tool with powerful public health applications, such as monitoring water quality. |
| - | Another interesting direction would be the linking of the [ | + | Another interesting direction would be the linking of the [https://2006.igem.org/UT_Austin_2005 light-sensing system] developed by the UT Austin iGEM team with electrical output in ''S. oneidensis''. In response to variations in light, the amount of electricity produced by ''S. oneidensis'' would change. This would allow for the intriguing possibility of not only ''S. oneidensis'' conveying information to the computer, but also the computer responding to the ''S. oneidensis''. A simple example would be that in response to a chemical input, ''S. oneidensis'' may increase its electrical output. Sensing this increase, the computer could turn on or off a light directed at the ''S. oneidensis'', modifying ''S. oneidensis'''s output, creating interesting feedback loops. This could ultimately be developed into more complex communications systems between bacteria and computers. We tried constructing this system over summer, but as the process requires making an EnvZ knockout strain of ''S. oneidensis'', we could not finish it. We did, however, make a few parts to facilitate future attempts. |
The possibilities are further broadened by our observations of co-cultures of ''E. coli'' and ''S. oneidensis''. Either of the systems described above could be pursued through an alternative alternative strategy of co-cultures. For instance, an array of ''E. coli'' which respond to different chemicals by breaking down lactose into lactate could be cultured with ''S. oneidensis''. In response to an increase in lactate, ''S. oneidensis'' would begin to produce higher levels of electricity. Co-cultures could also allow for more complex bacteria-computer interactions. This strategy could enable the coupling of almost any ''E. coli'' ability to electrical output. | The possibilities are further broadened by our observations of co-cultures of ''E. coli'' and ''S. oneidensis''. Either of the systems described above could be pursued through an alternative alternative strategy of co-cultures. For instance, an array of ''E. coli'' which respond to different chemicals by breaking down lactose into lactate could be cultured with ''S. oneidensis''. In response to an increase in lactate, ''S. oneidensis'' would begin to produce higher levels of electricity. Co-cultures could also allow for more complex bacteria-computer interactions. This strategy could enable the coupling of almost any ''E. coli'' ability to electrical output. | ||
These future directions in which our research can be taken demonstrate some of the exciting possibilities of BACTRICITY! | These future directions in which our research can be taken demonstrate some of the exciting possibilities of BACTRICITY! | ||
| - | |||
|} | |} | ||
| - | + | ||
<!--- end body ---> | <!--- end body ---> | ||
|} | |} | ||
Latest revision as of 05:03, 30 October 2008
|
|
 "
"