Team:Hawaii/Notebook/2008-08- 5
From 2008.igem.org
(Difference between revisions)
(→Discussion) |
(→Construction of p+r and g+t, otra vez) |
||
| (11 intermediate revisions not shown) | |||
| Line 10: | Line 10: | ||
===PCR contamination test (cont. from 8/2 and 8/4)=== | ===PCR contamination test (cont. from 8/2 and 8/4)=== | ||
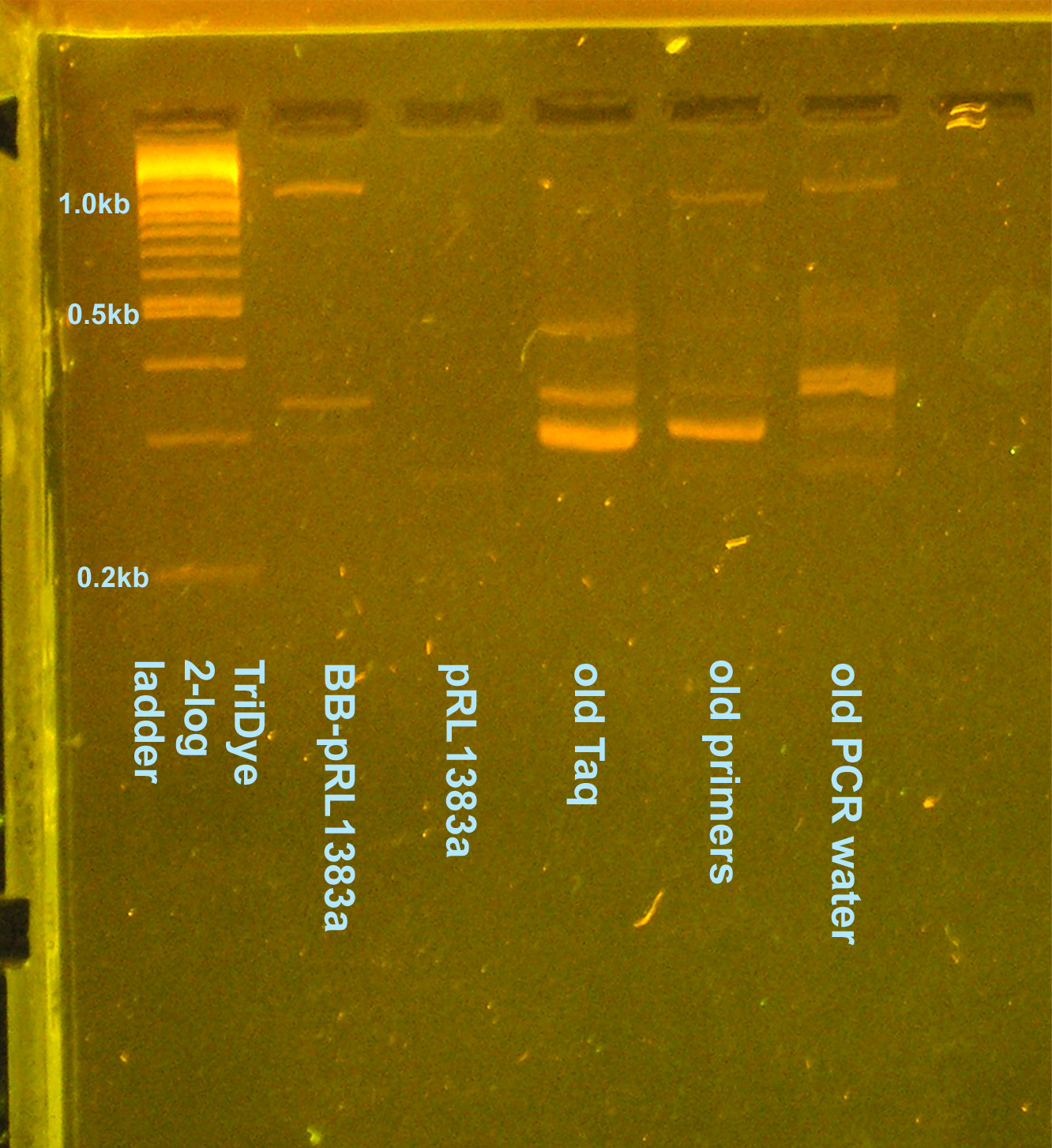
| + | [[Image:080508contaminationreretest.jpg|right|thumb|200px|EtBr stained 2.5% agarose gel ran at 95V for 1 hour. Ten microliters of the PCR reactions were loaded inot each well.]] | ||
:<strong>Grace</strong> | :<strong>Grace</strong> | ||
:* Ran an EtBr stained 2.5% gel at 95V for 2 hours | :* Ran an EtBr stained 2.5% gel at 95V for 2 hours | ||
::* Why aren't I getting consistent results for any of the lanes? | ::* Why aren't I getting consistent results for any of the lanes? | ||
| + | ::* GP says not to worry about contaminants -- they're there, let's move on with life | ||
| + | ::* On 8/5 gel, potentially see desired 1.1kb band | ||
| + | :::* Cut out of gel, use as template for another PCR to verify it's what we want --OR-- plasmid prep colony (decided to plasmid prep since we need to send in a plasmid prep to partsregistry eventually) | ||
| + | |||
| + | ===Inoculated for plasmid prep tomorrow === | ||
| + | :<strong>Grace</strong> | ||
| + | |||
| + | :* Colony #1 of pRL1383a 1:2 into LB+sp<sub>100</sub> | ||
| + | :* Jabbed nir and GFPf cryostocks and put into LB+amp<sub>100</sub> | ||
| + | ::* Also need plasmid prep of this to submit to registry | ||
| + | ::* Since both nir PCR RE'ds didn't show distinct bands, perhaps we should RE'd nir plasmid to make nir+rbs (if today's p+r doesn't work) | ||
===Construction of p+r and g+t, otra vez=== | ===Construction of p+r and g+t, otra vez=== | ||
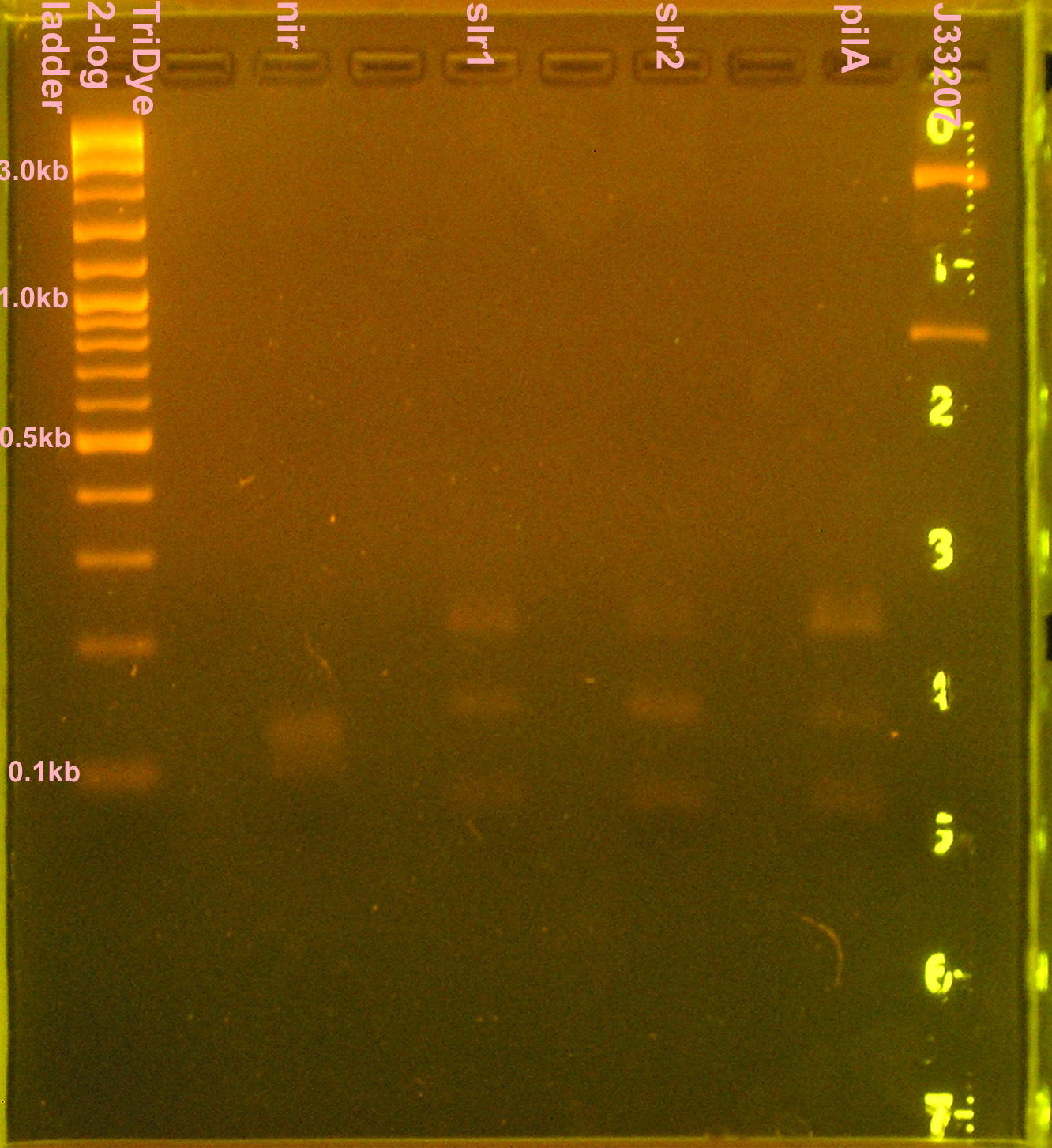
| + | [[Image:080508PCRRE.jpg|right|thumb|200px|EtBr stained 2% agarose gel ran at 60V for 2 hours. Twenty microliters of the restriction digests were loaded into each well.]] | ||
:<strong>Grace</strong> | :<strong>Grace</strong> | ||
:* 10 μl PCR reactions for nir, slr1, slr2, pilA, J33207 (in case transformation doesn't work) | :* 10 μl PCR reactions for nir, slr1, slr2, pilA, J33207 (in case transformation doesn't work) | ||
| - | :* RE digest of PCR products (no gel purification; used 3 μl PCR reactions) | + | :* RE digest of PCR products (no gel purification; used 3 μl PCR reactions) for 4 hours |
::* Digested nir and J33207 with EcoRI and SpeI | ::* Digested nir and J33207 with EcoRI and SpeI | ||
::* Digested slr1, slr2, pilA with XbaI and PstI | ::* Digested slr1, slr2, pilA with XbaI and PstI | ||
| - | :* Ran 20 μl of digests on 2% agarose gel stained with EtBr for | + | :* Ran 20 μl of digests on 2% agarose gel stained with EtBr for 2.5 hours |
| + | ::* Extracted slr1, slr2, pilA (bands <100bp) | ||
| + | ::* J33207 didn't cut? Bands at 830bp and 2.5kb | ||
| + | :::* VF-VR = 838; plasmid + part = ~2.7kb | ||
| + | ::* nir was a smear between 110-140bp; didn't cut because desired band (~90bp) not present | ||
| + | |||
| + | ===Re-streak=== | ||
| + | :<strong> Margaret</strong> | ||
| + | :* Restreaked stuff from [[Team:Hawaii/Notebook/2008-08- 4|yesterday]], including pSMC121 which was a lawn, the lac promoter (on fresh plate containing Kan50 & 1mM IPTG), and DB3.1 stock on fresh Sm50 plate. | ||
| + | |||
| + | ===Gels=== | ||
| + | :<strong> Margaret</strong> | ||
| + | |||
| + | :* ran gels for [[Team:Hawaii/Notebook/2008-08- 4|yesterday's]] PCR and Quantification experiments. | ||
| + | |||
| + | ===PCR=== | ||
| + | [[Image:aada_p1lytic.jpg|right|thumb|300px|PCR amplification of P1 lytic region, aadA from pRL1383a, and aadA from BioBrick Registry. Test.]] | ||
| + | :<strong> Margaret</strong> | ||
| + | |||
| + | :*testing PCR conditions for P1 lytic region and aadA (registry and pRL1383a) | ||
| + | |||
| + | <strong> Materials & Methods </strong> | ||
| + | |||
| + | :*Reaction conditions: 3.5ul water, 5ul taq, 0.5ul template (P1 lytic: pSB2K3(Plac), (+)&(-):pSB2K3(Plac): , aadA: pRL1383a, (+)&(-):pSB1A7, aadA: BioBrick Registry), 1ul primer (P1 lytic: p1lytic f&r, (+)&(-):VF2&VR , aadA: aadA f&r, (+)&(-):VF2&VR, aadA: aadA f&r) | ||
| + | |||
| + | :*Running Conditions: | ||
| + | ::*p1 lytic region: 94°C hold, 94°C 2', 95°C 30", <strong>54.2°C 30", 72°C 1'10"</strong>, 72°C 10', 4°C hold. | ||
| + | ::*aadA (pRL1383a & BB registry): 94°C hold, 94°C 2', 95°C 30", <strong>50°C 30", 72°C 30"</strong>, 72°C 10', 4°C hold. | ||
| + | |||
| + | <strong>Results</strong> | ||
| + | |||
| + | :* <strong>Lane 1</strong> contains the tri-dye 2-log ladder, which is not well resolved. K&G had better resolution of the ladder at lower voltage. <strong>Lane 2</strong> contains the amplification of the P1 lytic region from the I14032 prep which is on pSB2K3. This part did not amplify. The positive and negative controls in <strong>Lanes 3&4</strong> are actually the same thing: pSB2K3 (containing I14032) and VF2+VR. I mistakenly put Primer and Template in the negative. I do get amplification, though, which tells me the plasmid is there. The band should be around 353bp but looks to be above 400bp. This discrepancy is found in all amplifications of this part (which I have verified twice before: [[Team:Hawaii/Notebook/2008-08- 4|here]] and [[Team:Hawaii/Notebook/2008-07-30|here]]. K&G also amplified it directly from the BB registry and got the same band. <strong>Lane 5</strong> aadA from pRL1383a was amplified and large amounts of the right size are visualized on the gel. <strong>Lanes 6&7</strong> are the positive and negative control but this time VF2 and VR are amplifying pSB1A7 (proven on several occasions to amplify consistently; like [[Team:Hawaii/Notebook/2008-08- 4|here]]). The same mistake with the other controls in this experiment happended:(. Finally, <strong>lane 8</strong> contains the amplification of aadA from the BioBrick Registry. It has the right size and large amounts. | ||
== Drylab Work == | == Drylab Work == | ||
Latest revision as of 07:52, 7 August 2008
| Projects | Events | Resources | ||
|---|---|---|---|---|
| Sponsors | Experiments | Milestones | Protocols | |
| Notebook (t) | Meetings (t) |
Things we did today
Wetlab work
Transformation
- Grace
- Transformed 50 μl DB3.1 competent cells with 5 μl ligation reactions of p+r or g+t
- Incubated on ice 17 minutes after adding DNA
PCR contamination test (cont. from 8/2 and 8/4)
- Grace
- Ran an EtBr stained 2.5% gel at 95V for 2 hours
- Why aren't I getting consistent results for any of the lanes?
- GP says not to worry about contaminants -- they're there, let's move on with life
- On 8/5 gel, potentially see desired 1.1kb band
- Cut out of gel, use as template for another PCR to verify it's what we want --OR-- plasmid prep colony (decided to plasmid prep since we need to send in a plasmid prep to partsregistry eventually)
Inoculated for plasmid prep tomorrow
- Grace
- Colony #1 of pRL1383a 1:2 into LB+sp100
- Jabbed nir and GFPf cryostocks and put into LB+amp100
- Also need plasmid prep of this to submit to registry
- Since both nir PCR RE'ds didn't show distinct bands, perhaps we should RE'd nir plasmid to make nir+rbs (if today's p+r doesn't work)
Construction of p+r and g+t, otra vez
- Grace
- 10 μl PCR reactions for nir, slr1, slr2, pilA, J33207 (in case transformation doesn't work)
- RE digest of PCR products (no gel purification; used 3 μl PCR reactions) for 4 hours
- Digested nir and J33207 with EcoRI and SpeI
- Digested slr1, slr2, pilA with XbaI and PstI
- Ran 20 μl of digests on 2% agarose gel stained with EtBr for 2.5 hours
- Extracted slr1, slr2, pilA (bands <100bp)
- J33207 didn't cut? Bands at 830bp and 2.5kb
- VF-VR = 838; plasmid + part = ~2.7kb
- nir was a smear between 110-140bp; didn't cut because desired band (~90bp) not present
Re-streak
- Margaret
- Restreaked stuff from yesterday, including pSMC121 which was a lawn, the lac promoter (on fresh plate containing Kan50 & 1mM IPTG), and DB3.1 stock on fresh Sm50 plate.
Gels
- Margaret
- ran gels for yesterday's PCR and Quantification experiments.
PCR
- Margaret
- testing PCR conditions for P1 lytic region and aadA (registry and pRL1383a)
Materials & Methods
- Reaction conditions: 3.5ul water, 5ul taq, 0.5ul template (P1 lytic: pSB2K3(Plac), (+)&(-):pSB2K3(Plac): , aadA: pRL1383a, (+)&(-):pSB1A7, aadA: BioBrick Registry), 1ul primer (P1 lytic: p1lytic f&r, (+)&(-):VF2&VR , aadA: aadA f&r, (+)&(-):VF2&VR, aadA: aadA f&r)
- Running Conditions:
- p1 lytic region: 94°C hold, 94°C 2', 95°C 30", 54.2°C 30", 72°C 1'10", 72°C 10', 4°C hold.
- aadA (pRL1383a & BB registry): 94°C hold, 94°C 2', 95°C 30", 50°C 30", 72°C 30", 72°C 10', 4°C hold.
Results
- Lane 1 contains the tri-dye 2-log ladder, which is not well resolved. K&G had better resolution of the ladder at lower voltage. Lane 2 contains the amplification of the P1 lytic region from the I14032 prep which is on pSB2K3. This part did not amplify. The positive and negative controls in Lanes 3&4 are actually the same thing: pSB2K3 (containing I14032) and VF2+VR. I mistakenly put Primer and Template in the negative. I do get amplification, though, which tells me the plasmid is there. The band should be around 353bp but looks to be above 400bp. This discrepancy is found in all amplifications of this part (which I have verified twice before: here and here. K&G also amplified it directly from the BB registry and got the same band. Lane 5 aadA from pRL1383a was amplified and large amounts of the right size are visualized on the gel. Lanes 6&7 are the positive and negative control but this time VF2 and VR are amplifying pSB1A7 (proven on several occasions to amplify consistently; like here). The same mistake with the other controls in this experiment happended:(. Finally, lane 8 contains the amplification of aadA from the BioBrick Registry. It has the right size and large amounts.
Drylab Work
Sequencing
- Grace
- Reviewed sequencing results from 7/21, emailed CORE Hawaii with concerns
- Mislabeled samples
- Poor quality reads
Discussion
- Looks like our TriDye 2-log ladder has gone bad (500bp band splits into two bands). Norman will aliquot out new ladder.
Quote of the Day
History is the only laboratory we have in which to test the consequences of thought. - Étienne Gilson
[http://manoa.hawaii.edu/  ][http://manoa.hawaii.edu/ovcrge/
][http://manoa.hawaii.edu/ovcrge/  ][http://www.ctahr.hawaii.edu
][http://www.ctahr.hawaii.edu  ]
]
 "
"