Team:Hawaii/Notebook/2008-08- 8
From 2008.igem.org
(Difference between revisions)
(→Prep for sequencing) |
(→Construct p+r+g and p+r+s) |
||
| (9 intermediate revisions not shown) | |||
| Line 5: | Line 5: | ||
===Restreaked nir+rbs and I14032+rbs constructs=== | ===Restreaked nir+rbs and I14032+rbs constructs=== | ||
:<strong> Grace</strong> | :<strong> Grace</strong> | ||
| + | |||
| + | ===Plasmid prep (finished up)=== | ||
| + | :<strong>Grace</strong> | ||
| + | |||
| + | :* Resuspended plasmid preps in 50 μl TE buffer | ||
| + | :* Determined DNA concentrations of plasmids | ||
| + | {|class=wikitable border=1 align=center | ||
| + | !Plasmid | ||
| + | ! DNA Concentration | ||
| + | |- | ||
| + | |align=center |nir | ||
| + | |align=center |496.3 ng/μl | ||
| + | |- | ||
| + | |align=center |GFPf | ||
| + | |align=center |484.8 ng/μl | ||
| + | |- | ||
| + | |align=center |BB-pRL1383a | ||
| + | |align=center |508.0 ng/μl | ||
| + | |} | ||
===Prep for sequencing=== | ===Prep for sequencing=== | ||
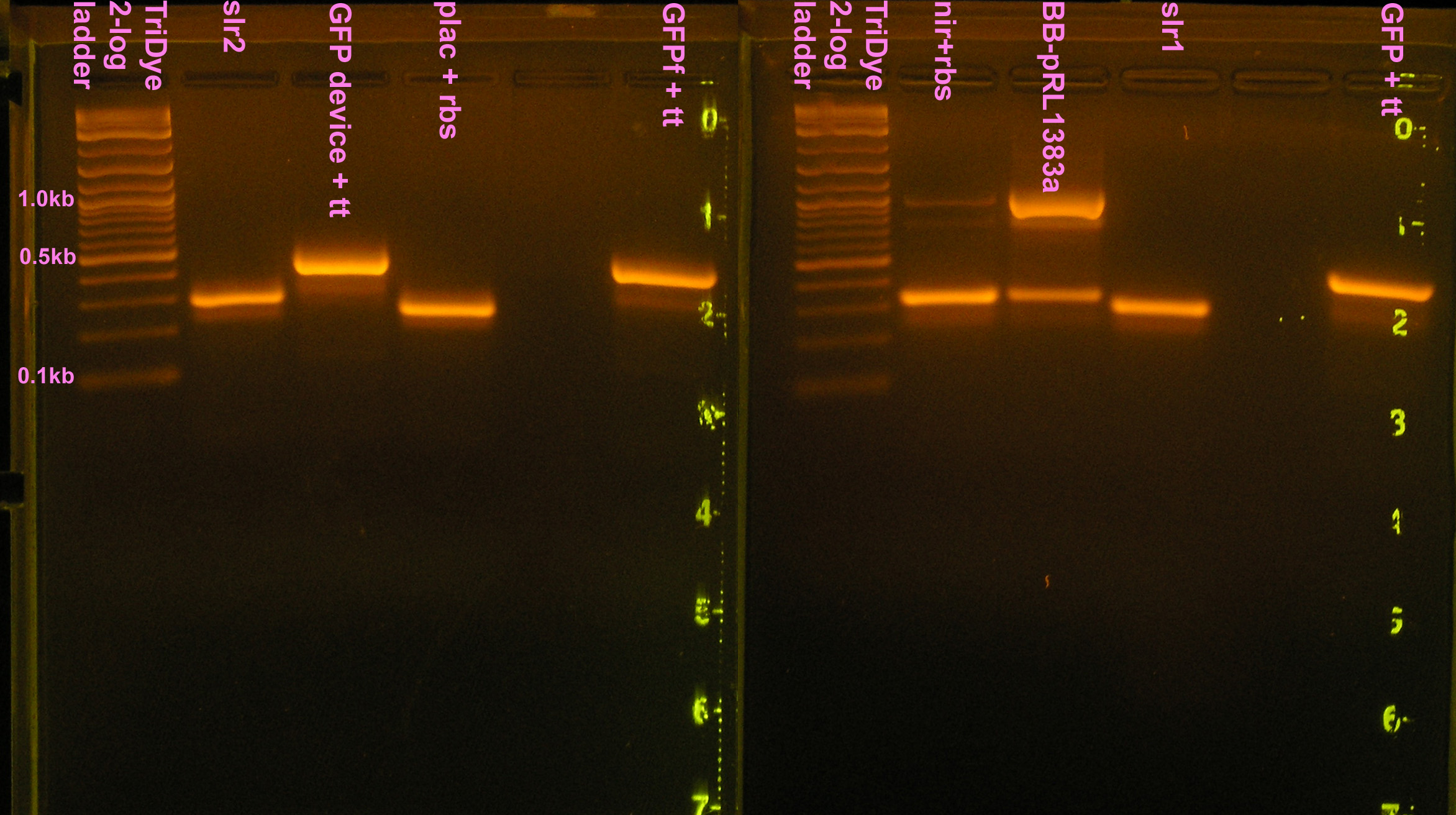
| - | [[Image: | + | [[Image:080808seqPCR.jpg|right|thumb|400px|EtBr stained 4% agarose gel ran at 60V for 100 min. Twenty-five microliters of the PCR reactions were loaded into each well.]] |
:<strong>Grace</strong> | :<strong>Grace</strong> | ||
:* 25 μl PCR reactions of nir+rbs, I14032+rbs, slr1, slr2, BB-pRL1383a | :* 25 μl PCR reactions of nir+rbs, I14032+rbs, slr1, slr2, BB-pRL1383a | ||
| Line 13: | Line 32: | ||
:* 25 μl PCR reactions of GFP+tt, GFPf+tt, J33207+tt | :* 25 μl PCR reactions of GFP+tt, GFPf+tt, J33207+tt | ||
::* Colony PCR indicates no ligation. Picked new colony, sequence to confirm failure. | ::* Colony PCR indicates no ligation. Picked new colony, sequence to confirm failure. | ||
| + | :* PCR of nir, B0015, B0030, B0034 for resequencing (bad reads last time) | ||
:* Gel purified all PCR rxns (we still have a problem with contaminant DNA/shadow bands) and desired bands were extracted from gel | :* Gel purified all PCR rxns (we still have a problem with contaminant DNA/shadow bands) and desired bands were extracted from gel | ||
::* 2% agarose gel ran at 60v for 2 hours | ::* 2% agarose gel ran at 60v for 2 hours | ||
:* Determined DNA concentrations via nanodrop spectrometer | :* Determined DNA concentrations via nanodrop spectrometer | ||
:* Prepared samples and sent to CORE Hawaii for sequencing | :* Prepared samples and sent to CORE Hawaii for sequencing | ||
| + | |||
| + | ===Construct p+r+g and p+r+s=== | ||
| + | :<strong>Krystle</strong> | ||
| + | [[Image:080808resdig.jpg|right|thumb|200px|EtBr stained 2% agarose gel ran at 60V for 60 min. Forty microliters of each digest were loaded.]] | ||
| + | :* Restriction Digest | ||
| + | :**nir+B0030, I14032+B0030, J33207 digested with SpeI and PstI | ||
| + | :**gfp, gfp''fusion'', and B0015 digested with XbaI and PstI | ||
| + | :* Gel Purified restriction digest | ||
| + | :** 2% agarose gel ran at 60 volts for 1.5 hours | ||
| + | ::: 40ul of the total restriction digest loaded into each well | ||
| + | ::* gfp''fusion'' cut out from gel | ||
= Discussion = | = Discussion = | ||
Latest revision as of 19:16, 9 August 2008
| Projects | Events | Resources | ||
|---|---|---|---|---|
| Sponsors | Experiments | Milestones | Protocols | |
| Notebook (t) | Meetings (t) |
Things we did today
Wetlab work
Restreaked nir+rbs and I14032+rbs constructs
- Grace
Plasmid prep (finished up)
- Grace
- Resuspended plasmid preps in 50 μl TE buffer
- Determined DNA concentrations of plasmids
| Plasmid | DNA Concentration |
|---|---|
| nir | 496.3 ng/μl |
| GFPf | 484.8 ng/μl |
| BB-pRL1383a | 508.0 ng/μl |
Prep for sequencing
- Grace
- 25 μl PCR reactions of nir+rbs, I14032+rbs, slr1, slr2, BB-pRL1383a
- Colony PCRs seem to indicate success
- 25 μl PCR reactions of GFP+tt, GFPf+tt, J33207+tt
- Colony PCR indicates no ligation. Picked new colony, sequence to confirm failure.
- PCR of nir, B0015, B0030, B0034 for resequencing (bad reads last time)
- Gel purified all PCR rxns (we still have a problem with contaminant DNA/shadow bands) and desired bands were extracted from gel
- 2% agarose gel ran at 60v for 2 hours
- Determined DNA concentrations via nanodrop spectrometer
- Prepared samples and sent to CORE Hawaii for sequencing
Construct p+r+g and p+r+s
- Krystle
File:080808resdig.jpg
EtBr stained 2% agarose gel ran at 60V for 60 min. Forty microliters of each digest were loaded.
- Restriction Digest
- nir+B0030, I14032+B0030, J33207 digested with SpeI and PstI
- gfp, gfpfusion, and B0015 digested with XbaI and PstI
- Gel Purified restriction digest
- 2% agarose gel ran at 60 volts for 1.5 hours
- 40ul of the total restriction digest loaded into each well
- gfpfusion cut out from gel
- Restriction Digest
Discussion
Quote of the Day
History is the only laboratory we have in which to test the consequences of thought. - Étienne Gilson
[http://manoa.hawaii.edu/  ][http://manoa.hawaii.edu/ovcrge/
][http://manoa.hawaii.edu/ovcrge/  ][http://www.ctahr.hawaii.edu
][http://www.ctahr.hawaii.edu  ]
]
 "
"