Team:Hawaii/Ligation of pRL1383a Parts
From 2008.igem.org
(Difference between revisions)
(picture 8/14) |
(explanations for 8-12) |
||
| Line 133: | Line 133: | ||
| | | | ||
|} | |} | ||
| + | |||
| + | <strong>Transformation & Verification PCR</strong> | ||
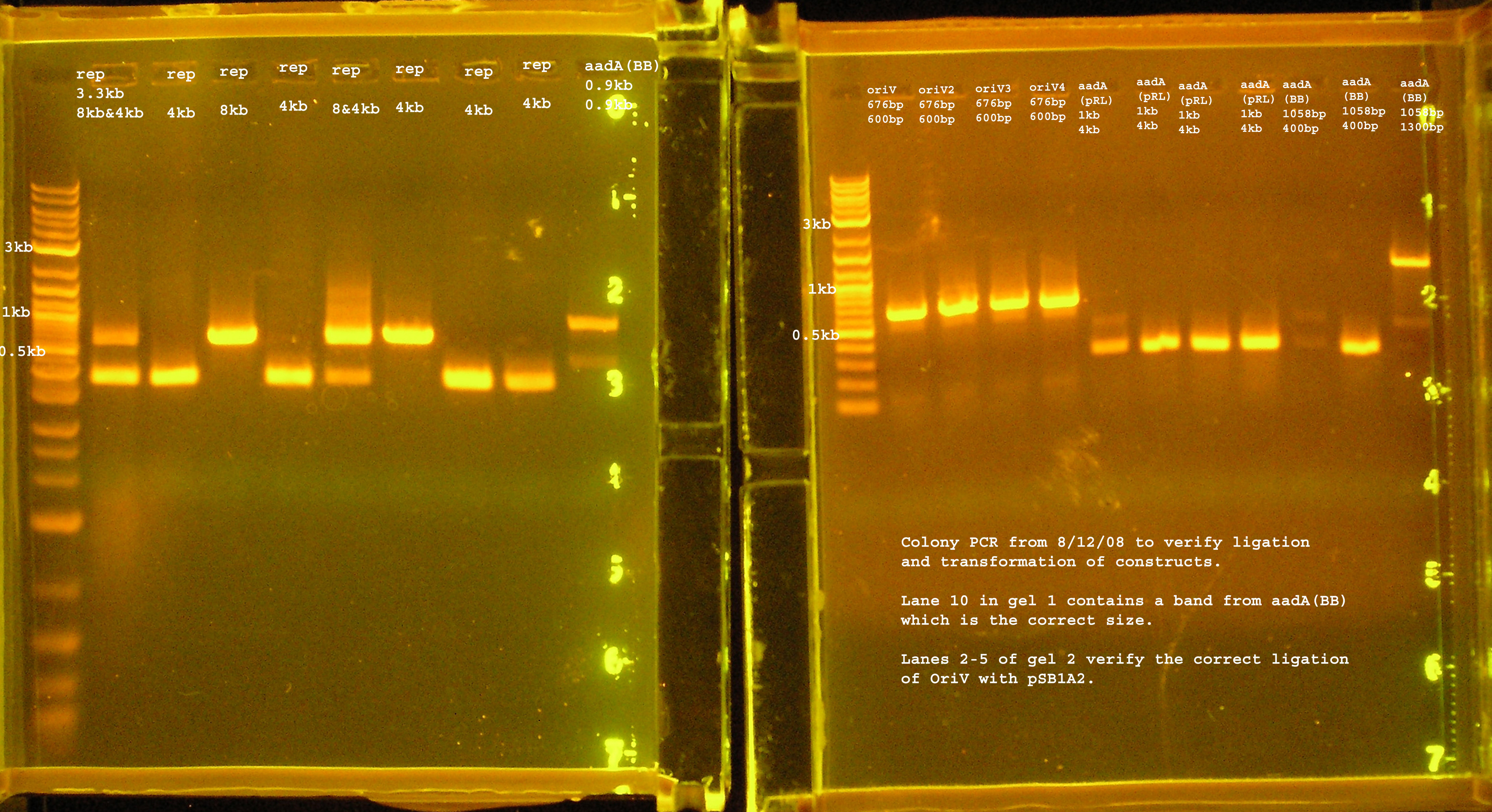
| + | [[Image:verification8_12_08.jpg|right|thumb|300px|PCR verification of colonies from transformation with plasmids containing rep, P1 lytic region, the aadA (BB&pRL1383a) region, and oriV.]] | ||
| + | |||
| + | :*The only bands corresponding to the correct size are the 4 oriVs and the 4th BB version of aadA. | ||
| + | :*The rep region lanes are all either 300 or 600bp when they should be 3.3kb. If the RBS (B0030) ligated back to itself, the band should be 253bp. The bands at 300bp may be explained by this. It is unclear what happened in the latter case. The incorrect aadA bands are also around 300bp which would mean the RBS containing vector ligated back to itself some how. B0030 was cut with SpeI and PstI. | ||
| + | |||
===8/14=== | ===8/14=== | ||
Revision as of 23:17, 21 August 2008
| Projects | Events | Resources | ||
|---|---|---|---|---|
| Sponsors | Experiments | Milestones | Protocols | |
| Notebook (t) | Meetings (t) |
Contents |
Ligation of Parts
- The BioBrick parts of pRL1383a are to be ligated in a series of experiments.
Methods
Restriction Digest
- Each part is digested with both enzymes. For all cases NEBuffer 2 is used.
- Reaction Conditions:
- 5ul Buffer
- 1ul Enzyme 1 + 1ul Enzyme 2
- 0.5 ul BSA
- Xul water
- Yul insert (1ug if possible)
- Zul vector (less than 1ug)
- Running Conditions: 2 hours at 37°C.
- Check the progress of the reaction by running a 0.8% gel.
Ligation
- Refer to http://openwetware.org/wiki/DNA_ligation for the reasoning behind this.
Transformation
Verification with Colony PCR
Results
7/27
- A few re-digests and ligations were performed today: oriT + pSB1A3, oriV + pSB1A3, mob + B0024, rep + R0010, aadA(pRL1383a+R0010). Afterwards, the ligation product was transformed to DH5-alpha. Only OriT transformed. I did not check the progress of this experiment with a gel, so I am not sure where this went wrong.
- Next time, a gel will be run after every step.
| Name | size | enzyme | quantity |
|---|---|---|---|
| oriT | ~125bp | XbaI & PstI | n/a |
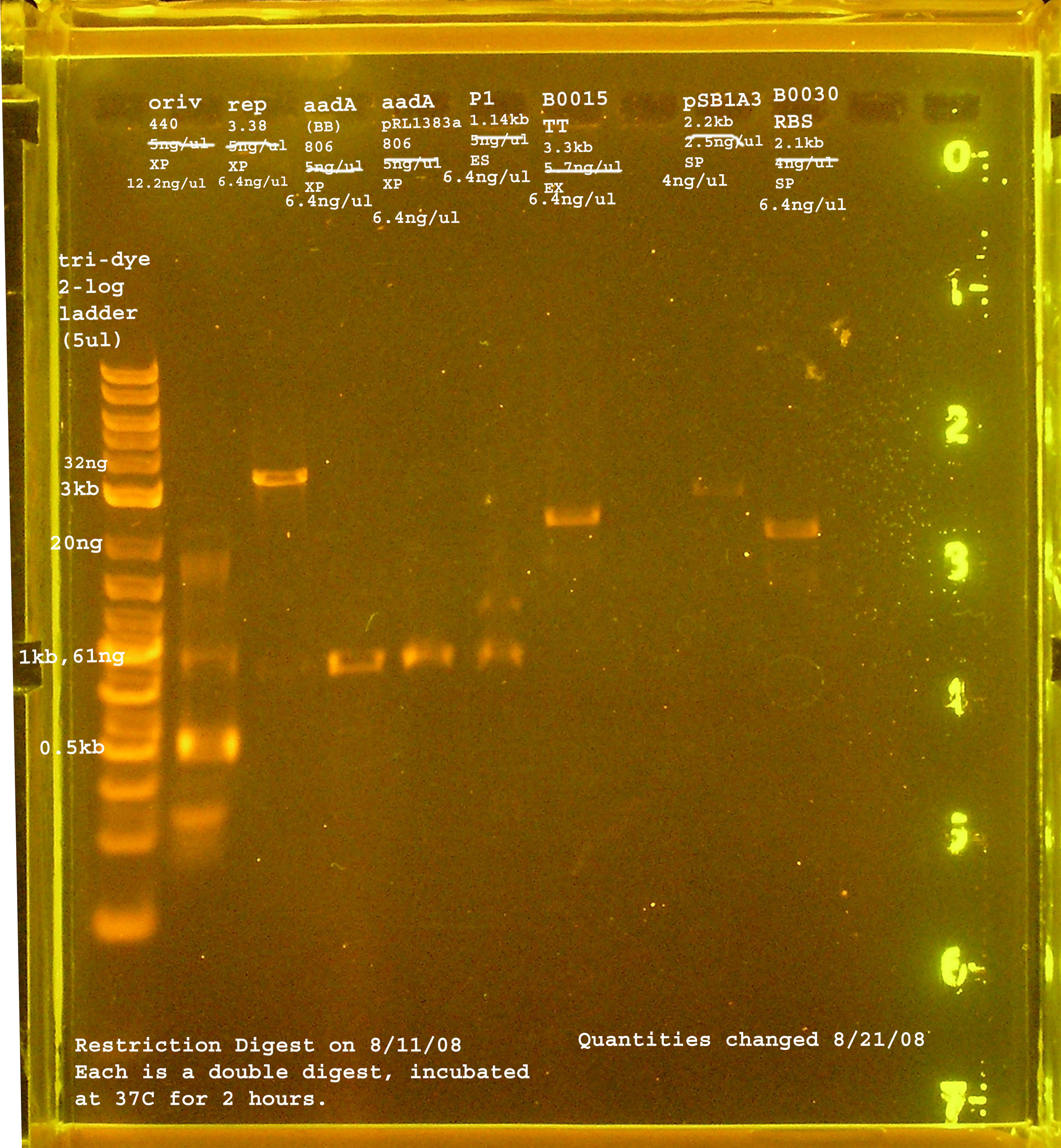
8/11
| Name | size | enzyme | quantity |
|---|---|---|---|
| rep | 3.3kb | XbaI & PstI | 5ng/ul |
| oriV | 415bp | XbaI & PstI | 5ng/ul |
| aadA (pRL1383a) | 806bp | XbaI & PstI | 5ng/ul |
| aadA (BB) | 806bp | XbaI & PstI | 5ng/ul |
| P1 lytic Region | 1.3kb | EcoRI & SpeI | 5ng/ul |
| [http://partsregistry.org/Part:BBa_B0030 BBa_B0030] | 2094bp | SpeI & PstI | 4ng/ul |
| [http://partsregistry.org/Part:BBa_B0015 BBa_B0015] | 3318bp | EcoRI & XbaI | 5.7ng/ul |
| [http://partsregistry.org/Part:pSB1A2 pSB1A2] | 2094bp | SpeI & PstI | 5.5ng/ul |
| Name | strain, antibiotics | colonies? after next day??? | PCR verification |
|---|---|---|---|
| rep(20ng)+B0030(16ng) | DH5-alpha & Amp100 | lawn | |
| oriV(20ng)+pSB1A2(50ng) | DH5-alpha & Amp100 | ||
| aadA (BB)+ B0030 | DH5-alpha & Amp100 | ||
| aadA(pRL1383a)+B0030 | DH5-alpha & Amp100 | ||
| P1 lytic + B0015 | DH5-alpha & Kan50 |
Transformation & Verification PCR
- The only bands corresponding to the correct size are the 4 oriVs and the 4th BB version of aadA.
- The rep region lanes are all either 300 or 600bp when they should be 3.3kb. If the RBS (B0030) ligated back to itself, the band should be 253bp. The bands at 300bp may be explained by this. It is unclear what happened in the latter case. The incorrect aadA bands are also around 300bp which would mean the RBS containing vector ligated back to itself some how. B0030 was cut with SpeI and PstI.
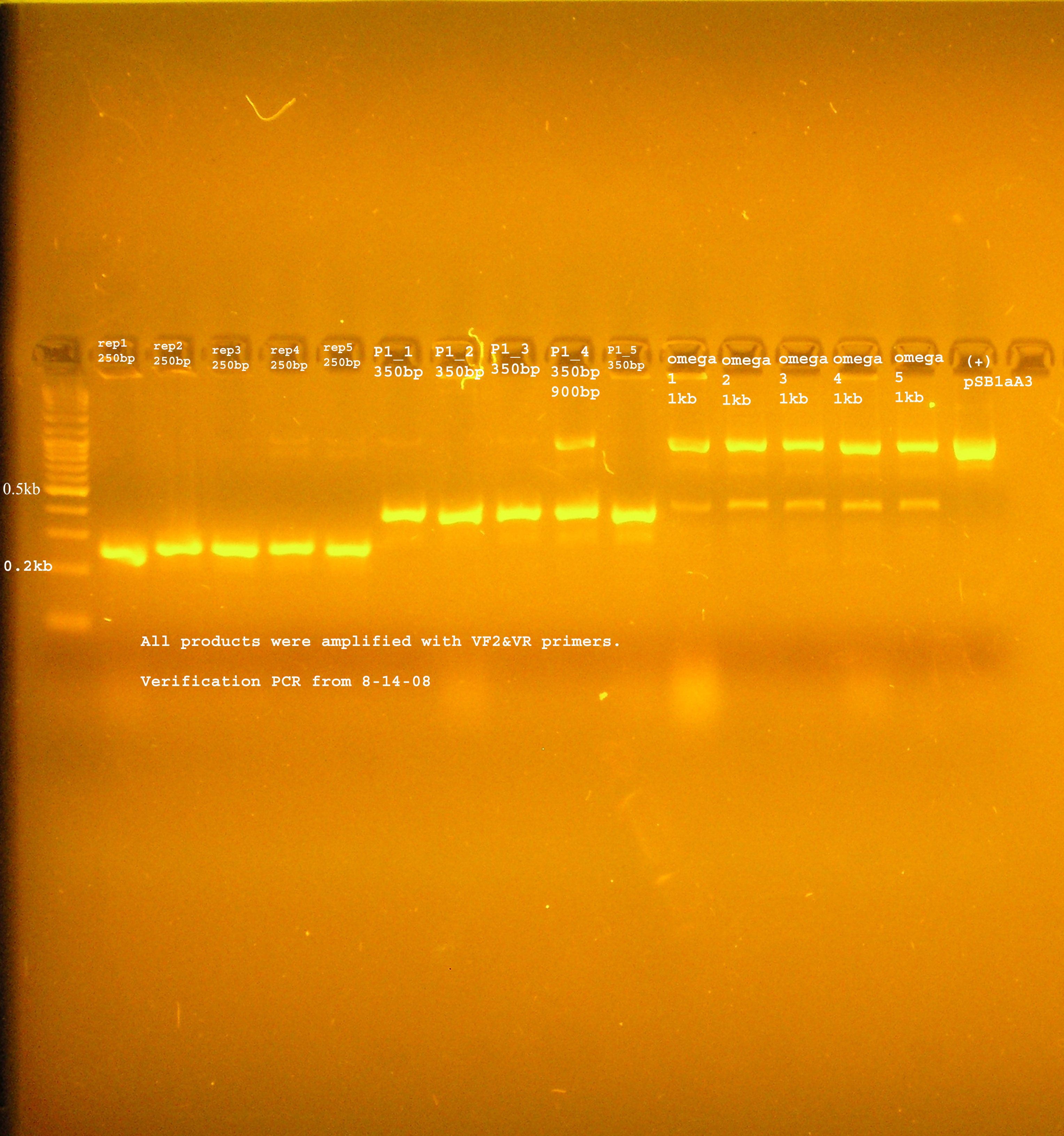
8/14
Discussion
- What was learned and how to do future experiments differently.
[http://manoa.hawaii.edu/  ][http://manoa.hawaii.edu/ovcrge/
][http://manoa.hawaii.edu/ovcrge/  ][http://www.ctahr.hawaii.edu
][http://www.ctahr.hawaii.edu  ]
]
 "
"