Team:Hawaii/Notebook/2008-08-27
From 2008.igem.org
(Difference between revisions)
(New page: {{Team:Hawaii/Header}} = Things we did today = == Wetlab work == ===Construction of p+r=== :<strong>Grace</strong> :* Ran RE digested products on gel :* Extracted nir, C0012 vector, BB-p...) |
(→Transformation) |
||
| (4 intermediate revisions not shown) | |||
| Line 4: | Line 4: | ||
== Wetlab work == | == Wetlab work == | ||
===Construction of p+r=== | ===Construction of p+r=== | ||
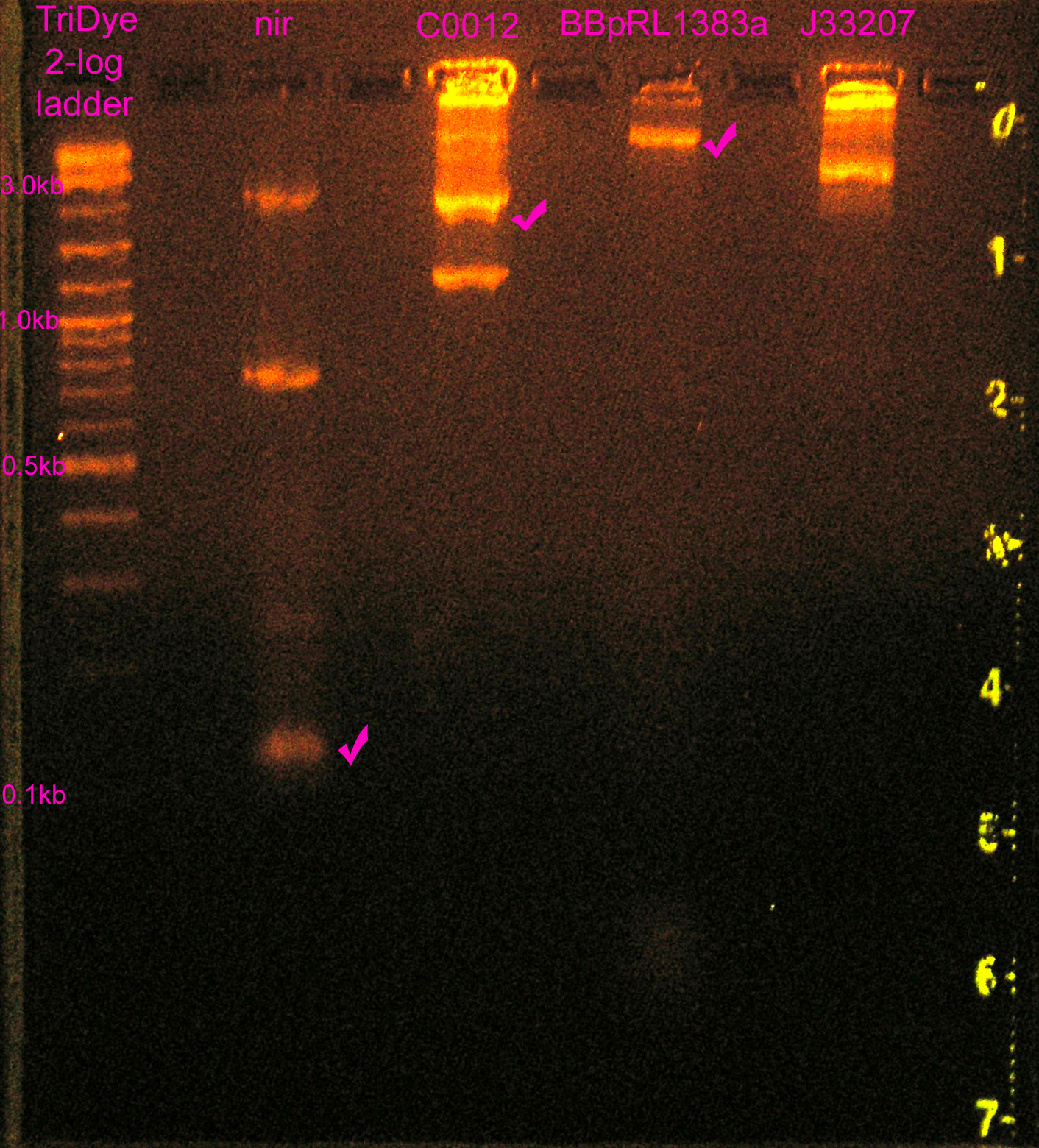
| + | [[Image:082708REdigests.jpg|right|thumb|250px|EtBr stained 2% agarose gel ran at 60V for 2.25 hours. Thirty microliters of RE digested DNA were loaded into each well.]] | ||
:<strong>Grace</strong> | :<strong>Grace</strong> | ||
:* Ran RE digested products on gel | :* Ran RE digested products on gel | ||
| + | ::* nir had three bands. Band at ~730bp = ? | ||
| + | ::* BB-pRL1383a -- still couldn't see cut out of GFP. Assumed double digest successful, too little DNA to see GFP. | ||
| + | ::* J33207 wrong size | ||
:* Extracted nir, C0012 vector, BB-pRL1383a from gel | :* Extracted nir, C0012 vector, BB-pRL1383a from gel | ||
:* 3A assembly (ligation) of C0012 vector (pSB1A2) with: | :* 3A assembly (ligation) of C0012 vector (pSB1A2) with: | ||
| Line 14: | Line 18: | ||
:* Transformed DB3.1 cells using 5 μl ligation reaction | :* Transformed DB3.1 cells using 5 μl ligation reaction | ||
| + | ===Transformation=== | ||
| + | :<strong>Margaret</strong> | ||
| + | :* ligation products from yesterday were transformed into DH5-a cells, pSMC121 was used as a positive control | ||
| + | :*rep+pSB1A3 (1:1) | ||
| + | :*rep+pSB1A3 (1:3) | ||
| + | :*rep+pSB1A3 (1:6) | ||
| + | :*P1 lytic +pSB1A3 (1:1) | ||
| + | :*P1 lytic +pSB1A3 (1:3) | ||
| + | :*P1 lytic +pSB1A3 (1:6) | ||
| + | :*[Plac B0030]+pSB1A3 (1:3) | ||
| + | :*[Plac B0030]+pSB1A3 (1:6) | ||
| + | :*[Plac B0034]+pSB1A3 (1:6)**ran out of vector | ||
| + | ===Sequencing=== | ||
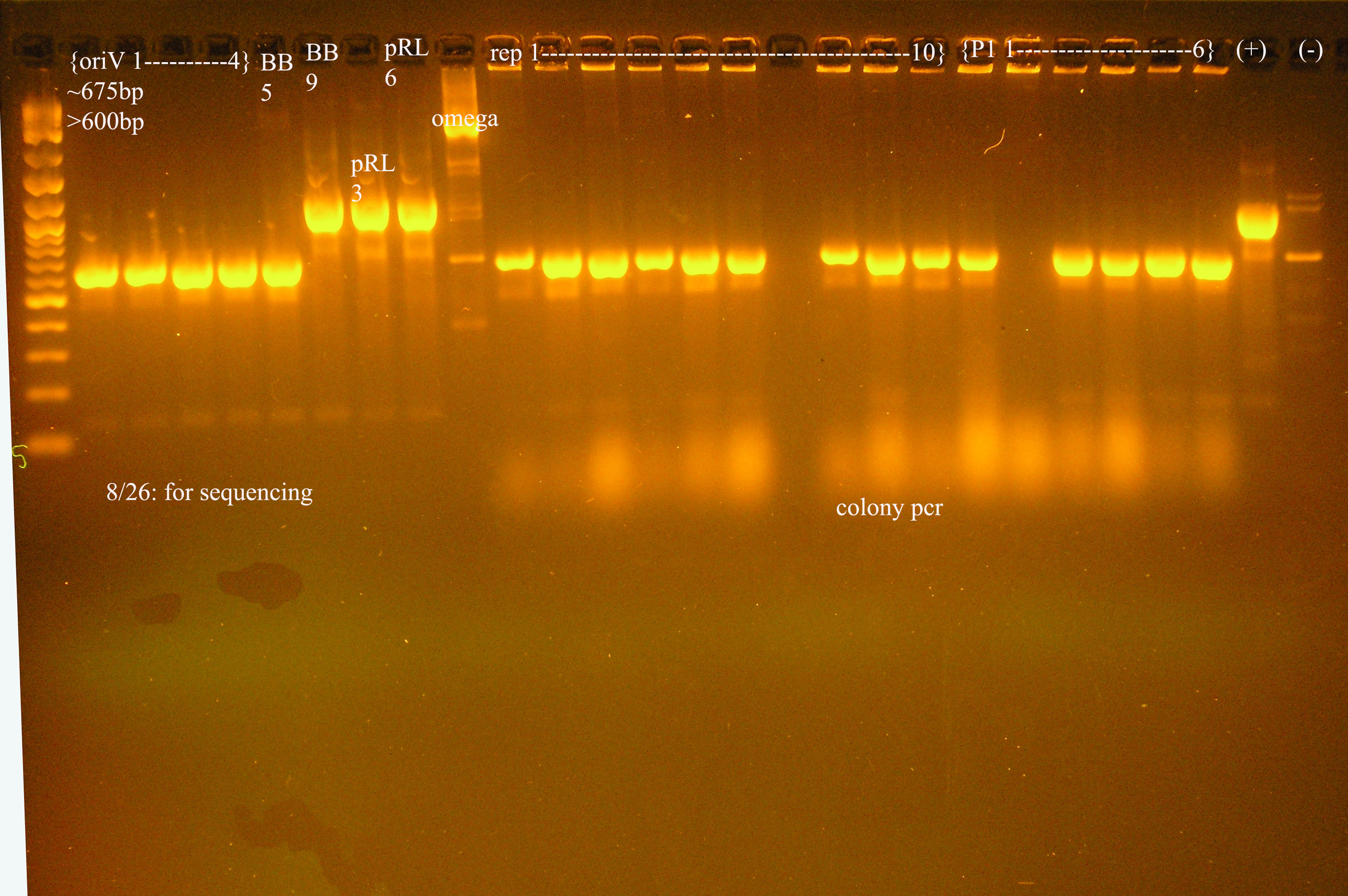
| + | [[Image:sequencing_gel.jpg|right|thumb|150px|Gel of the pcr in preparation for sequencing, also colony pcr of rep and lytic regions.]] | ||
| + | :<strong>Margaret</strong> | ||
| + | :*in preparation for sequencing, a PCR was run on the following plasmids: | ||
| + | oriV1-4, aadA(BB)5, aadA(BB)9, aadA(pRL)3, aadA(pRL)6, omega interposon | ||
| + | :*Note: In lane five, it looks like I added oriV instead of aada(BB)5. In the omega lane, there is a ladder. I will still sequence these. | ||
| + | :<strong>Grace</strong> | ||
| + | |||
| + | :* Prepared nir+rbs and rbs+GFP constructs for sequencing | ||
= Discussion = | = Discussion = | ||
Latest revision as of 08:28, 28 August 2008
| Projects | Events | Resources | ||
|---|---|---|---|---|
| Sponsors | Experiments | Milestones | Protocols | |
| Notebook (t) | Meetings (t) |
Things we did today
Wetlab work
Construction of p+r
- Grace
- Ran RE digested products on gel
- nir had three bands. Band at ~730bp = ?
- BB-pRL1383a -- still couldn't see cut out of GFP. Assumed double digest successful, too little DNA to see GFP.
- J33207 wrong size
- Extracted nir, C0012 vector, BB-pRL1383a from gel
- 3A assembly (ligation) of C0012 vector (pSB1A2) with:
- nir and rbs (B0034)
- plac and rbs(B0034)
- pSB1A2 (negative control)
- Transformed DB3.1 cells using 5 μl ligation reaction
Transformation
- Margaret
- ligation products from yesterday were transformed into DH5-a cells, pSMC121 was used as a positive control
- rep+pSB1A3 (1:1)
- rep+pSB1A3 (1:3)
- rep+pSB1A3 (1:6)
- P1 lytic +pSB1A3 (1:1)
- P1 lytic +pSB1A3 (1:3)
- P1 lytic +pSB1A3 (1:6)
- [Plac B0030]+pSB1A3 (1:3)
- [Plac B0030]+pSB1A3 (1:6)
- [Plac B0034]+pSB1A3 (1:6)**ran out of vector
Sequencing
- Margaret
- in preparation for sequencing, a PCR was run on the following plasmids:
oriV1-4, aadA(BB)5, aadA(BB)9, aadA(pRL)3, aadA(pRL)6, omega interposon
- Note: In lane five, it looks like I added oriV instead of aada(BB)5. In the omega lane, there is a ladder. I will still sequence these.
- Grace
- Prepared nir+rbs and rbs+GFP constructs for sequencing
Discussion
Quote of the Day
History is the only laboratory we have in which to test the consequences of thought. - Étienne Gilson
[http://manoa.hawaii.edu/  ][http://manoa.hawaii.edu/ovcrge/
][http://manoa.hawaii.edu/ovcrge/  ][http://www.ctahr.hawaii.edu
][http://www.ctahr.hawaii.edu  ]
]
 "
"