Team:Paris/Notebook/Protocols
From 2008.igem.org
(→Amplification of promoters) |
|||
| Line 96: | Line 96: | ||
= Purification (PROMEGA kit) = | = Purification (PROMEGA kit) = | ||
| - | + | == Gel Slice and PCR Product Preparation == | |
| - | + | === Dissolving the Gel Slice === | |
* Following electrophoresis, excise DNA band from gel slice in a pre-weighed 1.5 mL microcentrifuge tube. | * Following electrophoresis, excise DNA band from gel slice in a pre-weighed 1.5 mL microcentrifuge tube. | ||
* Add 10µL membrane Binding Solution per 10 mg of gel slice. We prefer not to vortex and we incubate at 50-65°C until gel slice is completely dissolved (∼10 min). Quick centrifuge. | * Add 10µL membrane Binding Solution per 10 mg of gel slice. We prefer not to vortex and we incubate at 50-65°C until gel slice is completely dissolved (∼10 min). Quick centrifuge. | ||
| - | + | === Processing PCR reactions === | |
For products above 40 pb | For products above 40 pb | ||
*Add an equal volume of Membrane Binding Solution to the PCR reaction. | *Add an equal volume of Membrane Binding Solution to the PCR reaction. | ||
| - | + | == Binding of DNA == | |
* Insert the SV Minicolumn into Collection Tube. | * Insert the SV Minicolumn into Collection Tube. | ||
* Transfer dissolved gel mixture or prepared PCR product to the Minicolumn assembly. Incubate at room temperature for 1 minute. | * Transfer dissolved gel mixture or prepared PCR product to the Minicolumn assembly. Incubate at room temperature for 1 minute. | ||
* Centrifuge at 16,000 x ''g'' for 1 minute. Discard the flowthrough and reinsert Minicolumn into Collection Tube. | * Centrifuge at 16,000 x ''g'' for 1 minute. Discard the flowthrough and reinsert Minicolumn into Collection Tube. | ||
| - | + | == Washing == | |
* Add 700 µL Membrane Wash Solution (ethanol added). Centrifuge at 16,000 x ''g'' for 1 minute. Discard flowthrough and reinsert the Minicolumn into Collection Tube. | * Add 700 µL Membrane Wash Solution (ethanol added). Centrifuge at 16,000 x ''g'' for 1 minute. Discard flowthrough and reinsert the Minicolumn into Collection Tube. | ||
* Repeat Step 4 with 500 µL Membrane Wash Solution. Centrifuge at 16,000 x ''g'' for 5 minutes. | * Repeat Step 4 with 500 µL Membrane Wash Solution. Centrifuge at 16,000 x ''g'' for 5 minutes. | ||
* Empty the Collection Tube and recentrifuge the column assembly for 1 min with the microcentrifuge lid open (or off) to allow evaporation of any residual ethanol. | * Empty the Collection Tube and recentrifuge the column assembly for 1 min with the microcentrifuge lid open (or off) to allow evaporation of any residual ethanol. | ||
| - | + | == Elution == | |
* Carefully transfer Minicolumn to a clean 1.5 mL microcentrifuge tube. | * Carefully transfer Minicolumn to a clean 1.5 mL microcentrifuge tube. | ||
* Add 30 µL Buffer EB (Qiagen). Incubate at room temparature for 1 minute. Centifuge at 16,000 x ''g'' for 1 minute. | * Add 30 µL Buffer EB (Qiagen). Incubate at room temparature for 1 minute. Centifuge at 16,000 x ''g'' for 1 minute. | ||
| Line 121: | Line 121: | ||
| - | + | = Quantification by Electrophoresis = | |
* Gel : 1.5-2% agarose with BET added (5 µL BET for 100 mL TBE) | * Gel : 1.5-2% agarose with BET added (5 µL BET for 100 mL TBE) | ||
* 10 µL Quick-Load 1 kb DNA Ladder | * 10 µL Quick-Load 1 kb DNA Ladder | ||
| Line 127: | Line 127: | ||
| - | + | = Ligation = | |
* 2 µL Ligase Buffer 10X | * 2 µL Ligase Buffer 10X | ||
* V<sub>v</sub> µL of measured [DNA]<sub>Vector</sub> in ng/µL (30 to 50 ng) | * V<sub>v</sub> µL of measured [DNA]<sub>Vector</sub> in ng/µL (30 to 50 ng) | ||
| Line 134: | Line 134: | ||
* 1 µL T4 DNA ligase at 400 000 U/mL concentration | * 1 µL T4 DNA ligase at 400 000 U/mL concentration | ||
* O/N at 16°C | * O/N at 16°C | ||
| - | |||
| - | |||
| - | |||
| - | ==Transformation | + | |
| + | ''V<sub>i</sub> = (3 x [DNA]<sub>Vector</sub> x V<sub>v</sub> x Lenght<sub>Insert</sub>) '''/''' ([DNA]<sub>Insert</sub> x Lenght<sub>Vector</sub>)'' | ||
| + | |||
| + | = Transformation = | ||
Use of TOP10 chemically competent cells | Use of TOP10 chemically competent cells | ||
| Line 154: | Line 154: | ||
* Incubate O/N at 37°C | * Incubate O/N at 37°C | ||
| - | + | = PCR Screening = | |
Use of 8 clones of Ligation transformants for screening PCR | Use of 8 clones of Ligation transformants for screening PCR | ||
| Line 181: | Line 181: | ||
| - | + | == Electrophoresis Purification of PCR == | |
* 10µl of ladder 100 pb or 1 kb | * 10µl of ladder 100 pb or 1 kb | ||
| Line 187: | Line 187: | ||
* Migration at 100V on '''1,5%''' gel until 3/4 way | * Migration at 100V on '''1,5%''' gel until 3/4 way | ||
| - | + | = Sequencing = | |
| - | + | [http://institut.cochin.inserm.fr/rubric_recherche/Plates-Formes/sequencage_genomique/I18NFolder.2005-02-10.4781618697/page2/fr Sequencing COCHIN] | |
| - | + | = Promoter Characterization Plan = | |
For theoretical consideration, see [[Team:Paris/Modeling/estimation|estimation of parameters]] | For theoretical consideration, see [[Team:Paris/Modeling/estimation|estimation of parameters]] | ||
| Line 420: | Line 420: | ||
|} | |} | ||
| - | + | = Protocol to make competent bacteria = | |
| + | 1. Use non competent bacteria (ex: MG1655) stocked in 1.5 mL LB (20% Glycerol): put a toothpick in the 1.5 mL stock tube and then place it in a 50 mL Falcon with 5 ml LB medium. Over Night culture at 37°C / 300 rpm | ||
| - | + | 2. 1/100 dilution in LB medium QSP 50 mL in an erlenmeyer of 250 mL | |
| - | + | ||
| - | + | 3. Culture at 37°C / 300 rpm untill OD<sub>600</sub> reach 0.6 | |
| - | + | ||
| + | 4. Fast cooling at +4°C by gently shaking the erlen in ice | ||
---- | ---- | ||
| Line 436: | Line 438: | ||
---- | ---- | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | 5. Use pre-cooled centrifuge at +4°C. Centrifuge 50 mL of the culture in 50 mL falcon: +4°C / 5 min / 5000 rpm | |
| + | |||
| + | 6. Discard supernatant by inverting the tube, and resuspend the pellet with 1 mL of cold CaCl<sub>2</sub> and mix gently the suspension by up and down | ||
| + | |||
| + | 7. Add cold CaCl<sub>2</sub> QSP 20 mL and incubate 30 min / +4°C | ||
| + | |||
| + | 8. Centrifuge the suspension : +4°C / 5 min / 5000 rpm | ||
| + | |||
| + | 9. Discard supernatant by inverting the tube, and resuspend the pellet with 1 mL of cold CaCl<sub>2</sub> and mix gently the suspension by up and down | ||
| + | |||
| + | 10. Pre-warm the bain-marie at 42°C | ||
| + | |||
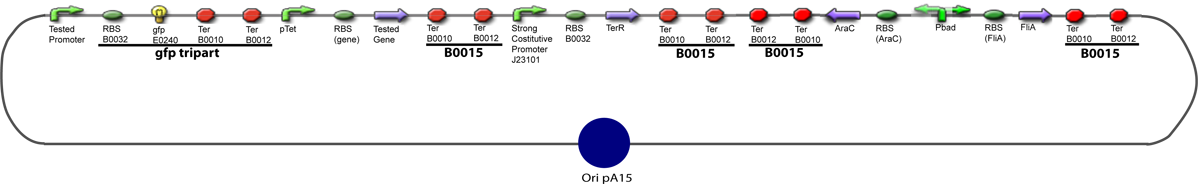
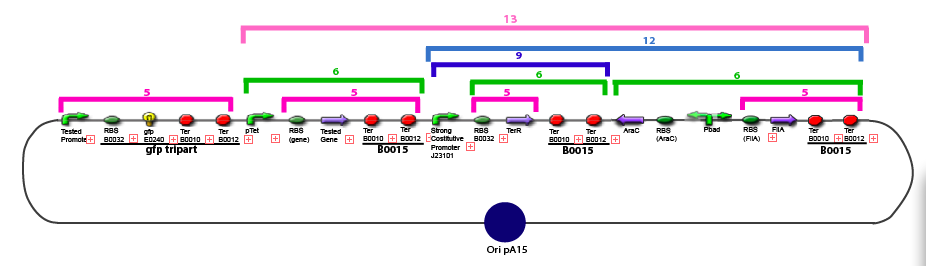
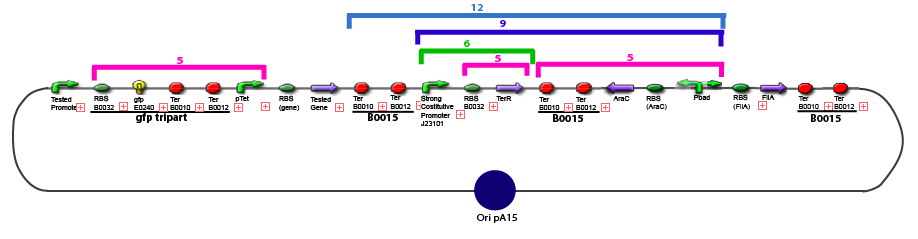
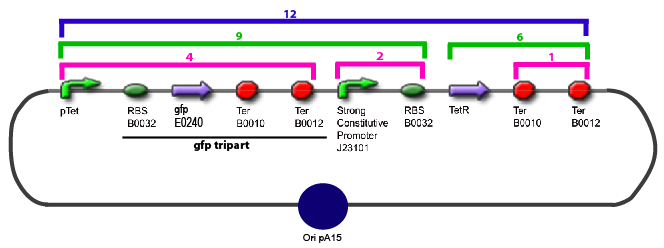
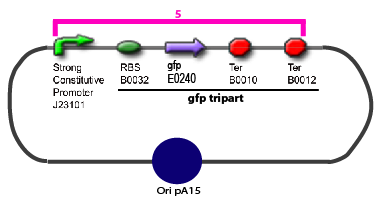
| + | 11. Add 2 µL of 1/100 diluted plasmid (containing a strong constitutive promoter [http://partsregistry.org/Part:BBa_J23101 J23101] before the [http://partsregistry.org/Part:BBa_E0240 GFP tripart]) to 100 µL of competent bacteria in a 15 ml falcon | ||
| + | |||
| + | 12. Incubate 20 min at +4°C | ||
| + | |||
| + | 13. Heat-Shock: 40 sec at 42°C / then 1 min in ice | ||
| + | |||
| + | 14. Add to the sample 1 mL LB medium without antibiotic and incubate 1h at 37°C / 300 rpm | ||
| + | |||
| + | 15. Spread 10 to 100 µL on plates with LB medium and the appropriate antibiotic | ||
| + | |||
| + | 16. Incubate O/N at 37°C / 300 rpm | ||
| + | |||
| + | 17. Prepare a [https://2008.igem.org/Team:Paris/Notebook/Protocols#Glycerol_Stocks Glycerol Stock] or/and use the transformed bacteria to study [https://2008.igem.org/Team:Paris/Notebook/Protocols#Study_of_the_doubling_time_of_the_bacteria_population the doubling time of the bacteria population] | ||
| + | |||
| + | = Study of the doubling time of the bacteria population = | ||
*Medium: | *Medium: | ||
- B1 Vitamin 0.1% | - B1 Vitamin 0.1% | ||
Revision as of 11:50, 28 August 2008
Culture of Stable strain with biobricks 2008
Glycerol Stocks
Minipreps (QIAGEN kit)
ElectrophoresisAn electrophoresis can be done to check if there is Product of Miniprep
Concentration of the MiniprepBy biophotometry
Check if the ratio 260/280 is over 1,6 Think about the dilution ! Digestion
Migration after digestion
Separate each band by an empty one ! Extraction
Amplification of promoters(to amplify the sequence in order to have enough amount of DNA to carry out the following of our experiments)
For each samples
1 µl dNTP
LID : 105°C Purification (PROMEGA kit)Gel Slice and PCR Product PreparationDissolving the Gel Slice
Processing PCR reactionsFor products above 40 pb
Binding of DNA
Washing
Elution
Quantification by Electrophoresis
Ligation
TransformationUse of TOP10 chemically competent cells
PCR ScreeningUse of 8 clones of Ligation transformants for screening PCR
After, add
Store the tubes on ice waiting for PCR attains 95°C then put the tubes in the machine
LID 105°C
Electrophoresis Purification of PCR
Sequencing[http://institut.cochin.inserm.fr/rubric_recherche/Plates-Formes/sequencage_genomique/I18NFolder.2005-02-10.4781618697/page2/fr Sequencing COCHIN]
Promoter Characterization PlanFor theoretical consideration, see estimation of parameters
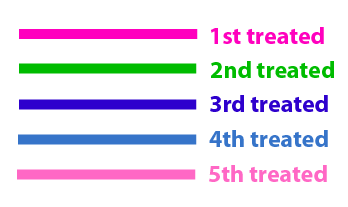
The same colour coded steps can be perfomed at the same time if elements needed are available The order for treating the colours should of course be:
This table contains the promoters we need to characterize, the transcription factors whose effect on the promoter we want to test, and the plasmid we want to obtain in order to carry out each characterization
Protocol to make competent bacteria1. Use non competent bacteria (ex: MG1655) stocked in 1.5 mL LB (20% Glycerol): put a toothpick in the 1.5 mL stock tube and then place it in a 50 mL Falcon with 5 ml LB medium. Over Night culture at 37°C / 300 rpm 2. 1/100 dilution in LB medium QSP 50 mL in an erlenmeyer of 250 mL 3. Culture at 37°C / 300 rpm untill OD600 reach 0.6 4. Fast cooling at +4°C by gently shaking the erlen in ice Before: prepare CaCl2 0.1M.
5. Use pre-cooled centrifuge at +4°C. Centrifuge 50 mL of the culture in 50 mL falcon: +4°C / 5 min / 5000 rpm 6. Discard supernatant by inverting the tube, and resuspend the pellet with 1 mL of cold CaCl2 and mix gently the suspension by up and down 7. Add cold CaCl2 QSP 20 mL and incubate 30 min / +4°C 8. Centrifuge the suspension : +4°C / 5 min / 5000 rpm 9. Discard supernatant by inverting the tube, and resuspend the pellet with 1 mL of cold CaCl2 and mix gently the suspension by up and down 10. Pre-warm the bain-marie at 42°C 11. Add 2 µL of 1/100 diluted plasmid (containing a strong constitutive promoter [http://partsregistry.org/Part:BBa_J23101 J23101] before the [http://partsregistry.org/Part:BBa_E0240 GFP tripart]) to 100 µL of competent bacteria in a 15 ml falcon 12. Incubate 20 min at +4°C 13. Heat-Shock: 40 sec at 42°C / then 1 min in ice 14. Add to the sample 1 mL LB medium without antibiotic and incubate 1h at 37°C / 300 rpm 15. Spread 10 to 100 µL on plates with LB medium and the appropriate antibiotic 16. Incubate O/N at 37°C / 300 rpm 17. Prepare a Glycerol Stock or/and use the transformed bacteria to study the doubling time of the bacteria population Study of the doubling time of the bacteria population
- B1 Vitamin 0.1%
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"