Team:BCCS-Bristol/Calendar-Notebook/15 July 2008
From 2008.igem.org
Rodgerread (Talk | contribs) |
Tgorochowski (Talk | contribs) |
||
| (5 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | <html><link rel="stylesheet" href="http:// | + | <html><link rel="stylesheet" href="http://homepage.mac.com/tgorochowski/iGEM/bccs-igem.css" type="text/css"></html> |
__NOTOC__ | __NOTOC__ | ||
<div class="bccsNavBar"> | <div class="bccsNavBar"> | ||
| Line 15: | Line 15: | ||
</div> | </div> | ||
| - | + | ==BioBrick Transformation== | |
| + | |||
| + | SOC medium was made in preparation for Biobrick transformation. This consisted of | ||
{| {{table}} | {| {{table}} | ||
| - | | align="center" style="background:#f0f0f0;"|''' | + | | align="center" style="background:#f0f0f0;"|'''Component''' |
| - | | align="center" style="background:#f0f0f0;"|''' | + | | align="center" style="background:#f0f0f0;"|'''Amount''' |
| + | |- | ||
| + | | Trypton||2g | ||
|- | |- | ||
| Yeast Extract||0.5g | | Yeast Extract||0.5g | ||
| Line 28: | Line 32: | ||
|} | |} | ||
| - | in 95ml of water, then autoclaved. | + | in 95ml of water, then autoclaved. After this the following was added: |
{| {{table}} | {| {{table}} | ||
| - | | align="center" style="background:#f0f0f0;"|''' | + | | align="center" style="background:#f0f0f0;"|'''Component''' |
| - | | align="center" style="background:#f0f0f0;"|''' | + | | align="center" style="background:#f0f0f0;"|'''Amount''' |
|- | |- | ||
| - | | Glucose||2ml | + | | MgCl<sub>2</sub> (2 M)||0.5ml |
| + | |- | ||
| + | | Glucose (1 M)||2ml | ||
|- | |- | ||
| | | | ||
|} | |} | ||
| - | + | The medium is stored at -20<sup>o</sup>C. This solution is from Molecular Cloning 3 A.2 SOB/SOC Medium. | |
| + | |||
| + | ==Chambers for bacteria-moving-bead assay== | ||
| + | |||
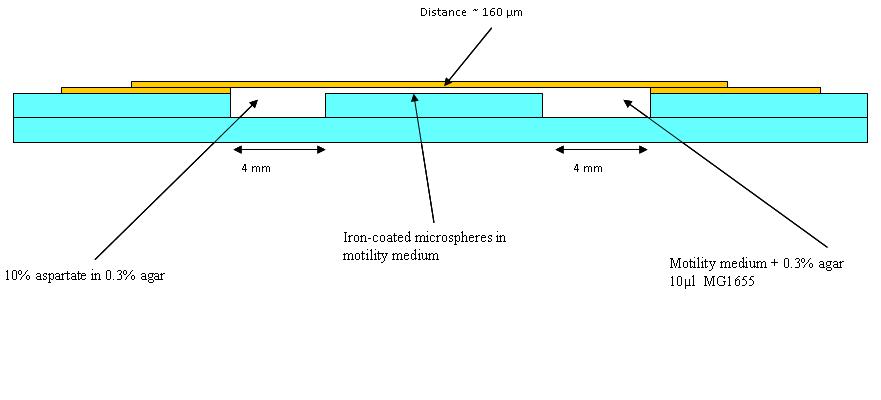
| + | The first trial in making wells as in the diagram (See below) was made, but it was found that a different glue is needed. Nevertheless, in the first chambers a blue dye in 0.3% agar (to represent aspartate) and an orange dye in 0.1% agar to represent the bacteria was put (beads were added in later experiments). Pictures were taken at 30 minutes intervals. This was to see how long a chemotatic gradient would take to set up. However, after three hours no gradient was obvious, so the chambers were left overnight. | ||
| + | |||
| + | [[Image:BCCS_Bristol-Wetlab-Well_diagram.JPG | 800px]] | ||
| + | |||
| + | ==Comparision of motility between ''E. coli'' MG1655 and MC1000== | ||
| + | |||
| + | The motility of both strains was tested under different conditions: | ||
| + | - agar concentration: 0.3 % and 0.5 % | ||
| - | + | - incubation temperatures: 25<sup>o</sup>C and 30<sup>o</sup>C | |
| - | + | The OD<sub>600</sub> was adjusted to 1.25 and the bacteria were inoculated in the middle of an LB-plate without yeast extract, but with streptomycin. After an incubation overnight there was no growth of MG1655 visible, because only MC1000 is resistant. Therefore, the experiment should be repeated. | |
Latest revision as of 09:53, 14 September 2008
BioBrick Transformation
SOC medium was made in preparation for Biobrick transformation. This consisted of
| Component | Amount |
| Trypton | 2g |
| Yeast Extract | 0.5g |
| NaCl | 0.05g |
in 95ml of water, then autoclaved. After this the following was added:
| Component | Amount |
| MgCl2 (2 M) | 0.5ml |
| Glucose (1 M) | 2ml |
The medium is stored at -20oC. This solution is from Molecular Cloning 3 A.2 SOB/SOC Medium.
Chambers for bacteria-moving-bead assay
The first trial in making wells as in the diagram (See below) was made, but it was found that a different glue is needed. Nevertheless, in the first chambers a blue dye in 0.3% agar (to represent aspartate) and an orange dye in 0.1% agar to represent the bacteria was put (beads were added in later experiments). Pictures were taken at 30 minutes intervals. This was to see how long a chemotatic gradient would take to set up. However, after three hours no gradient was obvious, so the chambers were left overnight.
Comparision of motility between E. coli MG1655 and MC1000
The motility of both strains was tested under different conditions:
- agar concentration: 0.3 % and 0.5 %
- incubation temperatures: 25oC and 30oC
The OD600 was adjusted to 1.25 and the bacteria were inoculated in the middle of an LB-plate without yeast extract, but with streptomycin. After an incubation overnight there was no growth of MG1655 visible, because only MC1000 is resistant. Therefore, the experiment should be repeated.
 "
"