Team:Valencia/Parts
From 2008.igem.org
| Line 1: | Line 1: | ||
| + | <html><link rel="stylesheet" href="https://2008.igem.org/wiki/index.php?title=User:Joadelas/valencia.css&action=raw&ctype=text/css" type="text/css"></html> | ||
| + | {{Valencia/Menu}} | ||
| + | |||
| + | |||
| + | <div class="valenciaMain"> | ||
| + | |||
| + | <div style=" width:94%; margin: 0 auto;"> | ||
| + | |||
'''Ucp1''' | '''Ucp1''' | ||
| Line 143: | Line 151: | ||
Y axis = Population in OD (optic density) units. | Y axis = Population in OD (optic density) units. | ||
X axis = Time in hours. | X axis = Time in hours. | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
Revision as of 13:20, 23 October 2008
Ucp1
General information
Ucp1 is a gene which encodes for UCP1 protein.
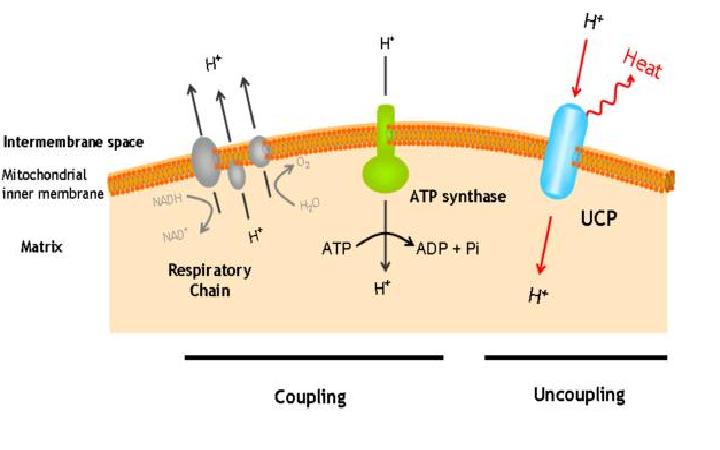
The uncoupling protein UCP1 is a proton carrier characteristic of brown adipose tissue. UCP1 uncouples the respiratory chain of ATP production, converting the metabolic energy in heat.
|
UCP1 is a 33kd protein which is exclusively located in the brown adipocites.
It is an integral protein present in inner mitocondrial membrane.
The protein has a tripartite structure. The structure displays an around 100 residues region which is three times repeated. Each part encodes for two transmembrane segments and one long hydrophilic loop.The functional carrier unit is an homodimer.
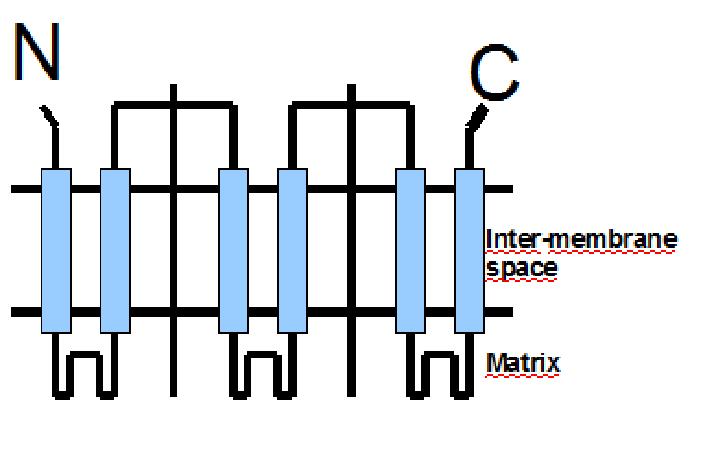
|
The main difference between UCP1 and most of the proteins with a nuclear codification is the lack of the importation targeting to the mitochondria in UCP+ proteins.
The condition that determines the mitochondria as the protein target lays in the first loop which protudes in the mitochondrial matrix.
The second loop of the matrix is essential for the insertion of the protein in the inner mitochondrial matrix. Purine nucleotides act as inhibitors of protein activity and esterificated fatty acids act as inductors.
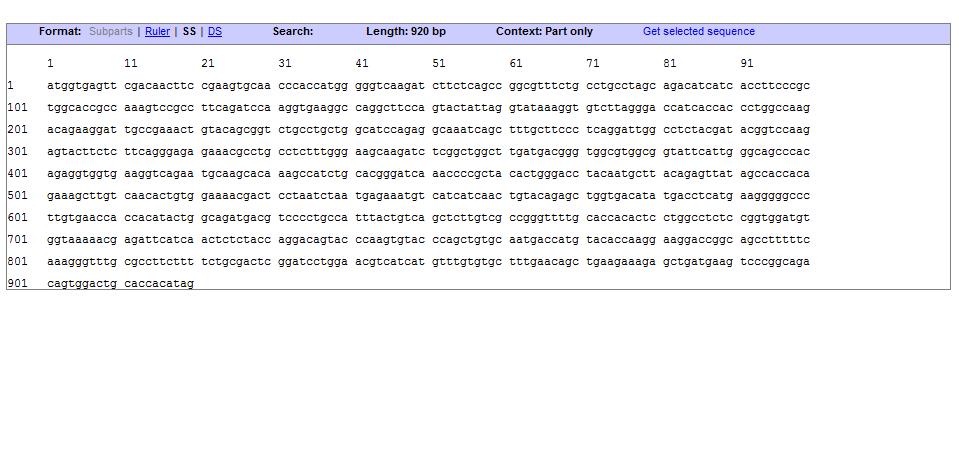
Sequence
UCP 175 deleted
The uncoupling protein UCP 175 deleted is a proton carrier which is obtaines from the direct mugenesis of the Ucp1 gene. Ucp 175 has a deletion in the triplet the encodes for the Glycine 175. The mutant shows a generation time and an heat up capacity higher than UCP1. UCP 175 deleted uncouples the respiratory chain of ATP production, converting the metabolic energy in heat. The mutant shows a generation time and an heat up capacity higher than UCP1.

|
UCP 175 deleted is a 33kd protein.
The protein has a tripartite structure. The structure displays an around 100 residues region which is three times repeated. Each part encodes for two transmembrane segments and one long hydrophilic loop.The functional carrier unit is an homodimer.

|
The main difference between UCP 175 deleted and most of the proteins with a nuclear codification is the lack of the importation targeting to the mitochondria in UCP 175 deleted proteins.
The condition that determines the mitochondria as the protein target lays in the first loop which protudes in the mitochondrial matrix.
The second loop of the matrix is essential for the insertion of the protein in the inner mitochondrial matrix.
That protein is without direct regulation and its generation time depends on the inner activity the protein.
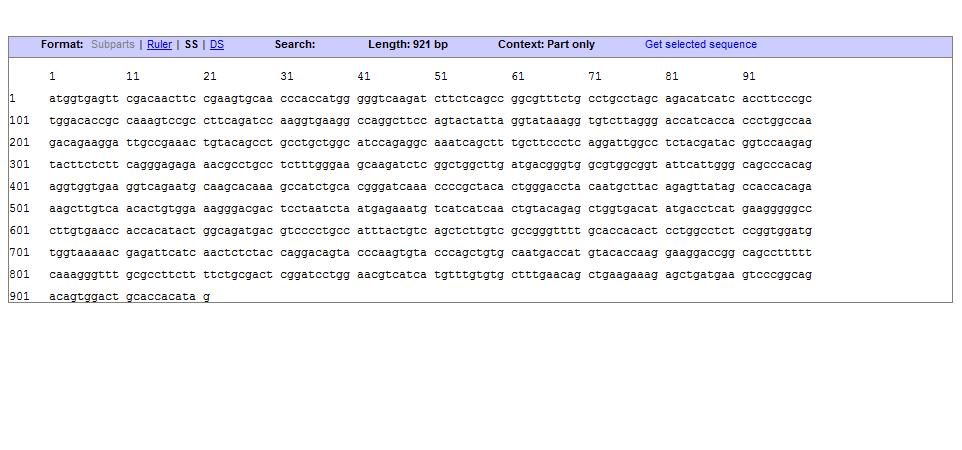
Sequence:
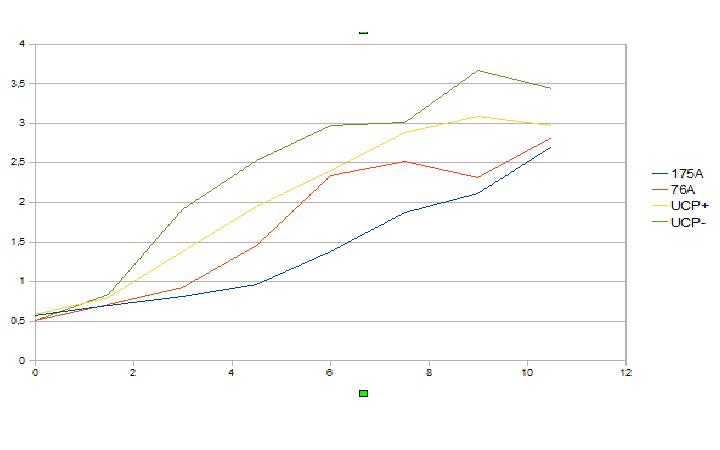
Graphic about the growth of four different strains of Saccharomyces cerevisiae transformed with four kinds of UCP genes:
Y axis = Population in OD (optic density) units. X axis = Time in hours.
UCP 76 deleted
The uncoupling protein UCP 76 deleted is a proton carrier which is obtaines from the direct mugenesis of the Ucp1 gene. Ucp 76 has a deletion in the triplet the encodes for the Glycine 76. The mutant shows a generation time and an heat up capacity higher than UCP1. UCP 76 deleted uncouples the respiratory chain of ATP production, converting the metabolic energy in heat. The mutant shows a generation time and an heat up capacity higher than UCP1.

|
UCP 76 deleted is a 33kd protein.
The protein has a tripartite structure. The structure displays an around 100 residues region which is three times repeated. Each part encodes for two transmembrane segments and one long hydrophilic loop.The functional carrier unit is an homodimer.

|
The main difference between UCP 76 deleted and most of the proteins with a nuclear codification is the lack of the importation targeting to the mitochondria in UCP 76 deleted proteins.
The condition that determines the mitochondria as the protein target lays in the first loop which protudes in the mitochondrial matrix.
The second loop of the matrix is essential for the insertion of the protein in the inner mitochondrial matrix.
That protein is without direct regulation and its generation time depends on the inner activity the protein.
Sequence:
Graphic about the growth of four different strains of Saccharomyces cerevisiae transformed with four kinds of UCP genes:
Y axis = Population in OD (optic density) units. X axis = Time in hours.
 "
"