Team:Heidelberg/Notebook/Killing II/3rdweek
From 2008.igem.org
(Difference between revisions)
(→Activity Test) |
(→Friday 08/22/2008) |
||
| (10 intermediate revisions not shown) | |||
| Line 482: | Line 482: | ||
| - | + | [[https://2008.igem.org/Team:Heidelberg/Notebook/Killing_II/3rdweek back]] | |
| - | + | ||
| - | + | ||
==Tuesday 08/19/2008== | ==Tuesday 08/19/2008== | ||
| Line 503: | Line 501: | ||
*Presentation in group seminar of Eils' group | *Presentation in group seminar of Eils' group | ||
| - | + | [[https://2008.igem.org/Team:Heidelberg/Notebook/Killing_II/3rdweek back]] | |
==Wednesday 08/20/2008== | ==Wednesday 08/20/2008== | ||
| Line 563: | Line 561: | ||
------- | ------- | ||
20.0 µl | 20.0 µl | ||
| - | *Transformation into ''E. coli'' TOP 10 cells (Transformation | + | *Transformation into ''E. coli'' TOP 10 cells (Transformation protocol Chris) |
*Cloning possibly not works because PstI cuts twice in pBAD18 | *Cloning possibly not works because PstI cuts twice in pBAD18 | ||
| + | |||
| + | [[https://2008.igem.org/Team:Heidelberg/Notebook/Killing_II/3rdweek back]] | ||
==Thursday 08/21/2008== | ==Thursday 08/21/2008== | ||
| Line 578: | Line 578: | ||
*Design of official Wiki | *Design of official Wiki | ||
| + | [[https://2008.igem.org/Team:Heidelberg/Notebook/Killing_II/3rdweek back]] | ||
==Friday 08/22/2008== | ==Friday 08/22/2008== | ||
| Line 590: | Line 591: | ||
*Cloning Results are tested via a activity test | *Cloning Results are tested via a activity test | ||
====Activity Test==== | ====Activity Test==== | ||
| - | *ONC was used for a sender activity test. The following graph shows the plate scheme for the test. (S = sender, R = Receiver, 1:5 = Sender:Receiver ratio 1:5 ...) [[Image:080822-platescheme_pBADsendertest.jpg | | + | *ONC was used for a sender activity test. The following graph shows the plate scheme for the test. (S = sender, R = Receiver, 1:5 = Sender:Receiver ratio 1:5 ...) [[Image:080822-platescheme_pBADsendertest.jpg | 374 px | center]] |
*Cells were incubated and GFP intensity as well as OD were measured every 2 hours. After 2 hours we obtained that we forget to induce LuxI production with arabinose. Because of that 2 µl of a 10% arabinose dilution was putted in every well. | *Cells were incubated and GFP intensity as well as OD were measured every 2 hours. After 2 hours we obtained that we forget to induce LuxI production with arabinose. Because of that 2 µl of a 10% arabinose dilution was putted in every well. | ||
| Line 611: | Line 612: | ||
**#Gelextraction of ligated receiver. | **#Gelextraction of ligated receiver. | ||
**#Ligation of pSB1A3 luxR-colicin-receiver. | **#Ligation of pSB1A3 luxR-colicin-receiver. | ||
| + | |||
| + | [[https://2008.igem.org/Team:Heidelberg/Notebook/Killing_II/3rdweek back]] | ||
| + | |||
[[Team:Heidelberg/Notebook/Killing_II/4thweek | go to 4<sup>th</sup> week]] | [[Team:Heidelberg/Notebook/Killing_II/4thweek | go to 4<sup>th</sup> week]] | ||
Latest revision as of 21:23, 28 October 2008


3rd week
Contents |
Monday 08/18/2008
Colicin Receiver
Cloning
- Literature research on different E. coli strains to see if they have BtuB receptor. This receptor is necessary for transport through the outer cellmembran in the prey cells. MG1655 and TOP 10 have these receptors. For DH5alpha we found no clear information.
Sender part
pBAD - Cloning
- Transformation of pBAD18 into TOP10 cells to amplify the plasmid.
- PCR of BBa_F1610 for pBAD18 cloning with pSB_ins Primer:
[back]
Tuesday 08/19/2008
Colicin Receiver
Cloning
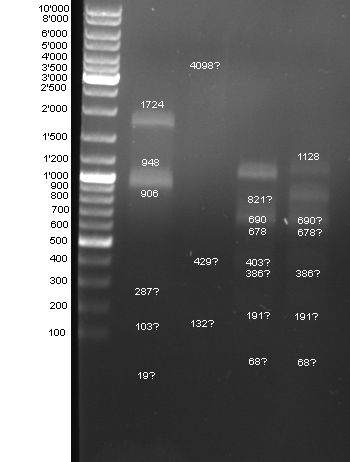
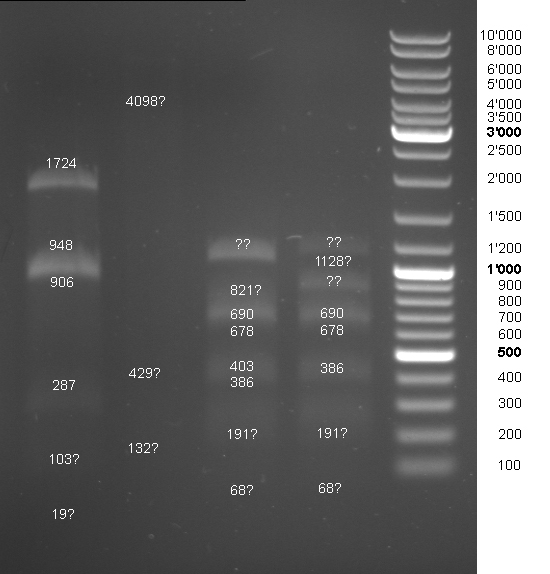
- Controldigestion of BBa_F2621, BBa_F2622, BBa_I15030, BBa_F1610 which received from the registry. See Wednesdaynesday 08/08/13 for protocoll: 1h 45 min
- Gel of Digestion:
- Results: To weak bands. New gel tomorrow.
- Inoculation of pColE9-J LB-liquid culture for maxiprep (250 ml)
Sender part
pBAD-Sender Cloning
- Inoculation of BBa_F1610 LB-liquid culture for maxiprep (250 ml)
- Inoculation of pBAD18 LB-liquid culture for miniprep (10 ml)
General
- Presentation in group seminar of Eils' group
[back]
Wednesday 08/20/2008
Colicin Receiver
Cloning
- Maxiprep of pColE9-J: CompactPrep Plasmid Mxi Kit 25, Qiagen
- Inoculation of liquid culture with pColE1 cells:
- Data of [http://www.dsmz.de/microorganisms/html/plasmids/plasmid.dsm003877.html DSMZ homepage]:
- Plasmid name: ColE1
- Other collection No: JC411
- Marker: E1imm
- DSMZ-Nr: 3877
- Molecular Weight: 6.6 kb
- Host:E. coli JC411
- Remarks: Naturally occurring plasmid; cells with plasmid ColE1 produce immunity proteins rendering the host immune to the lethal effects of the homologous colicin; non-transmissible; unique restriction site for EcoRI; non-conjugative; 10 - 15 copies per cell; amplifiable.
- Medium:
- Data of [http://www.dsmz.de/microorganisms/html/plasmids/plasmid.dsm003877.html DSMZ homepage]:
10.0 g Tryptone 5.0 g Yeast extract 10.0 g NaCl 1.0 L H2O +0.1 % Glucose
- Opening of the glass vial:
Double vial is closed under vacuum. Heat end of vial in a flame. Put 2-3 water drops on the spike to let the glass splitt. Use forceps to remove splitted end. Take out the inner vial. Remove the cotton plug and treat the inner side of the vial with flames. Put 0.5 ml of the media into the vial, and put the cotton plug back. Incubate 30 mins for 37 °C Stir solution with an inoculation loop an mix with 5 ml medium. Plate 100 µl of liquid culture on LB agarplate Incubate liquidculture and agarplate at 37 °C ON
- 2nd Gel of controldigestion see Tuesdaysday for details:
Sender part
pBAD-Sender Cloning
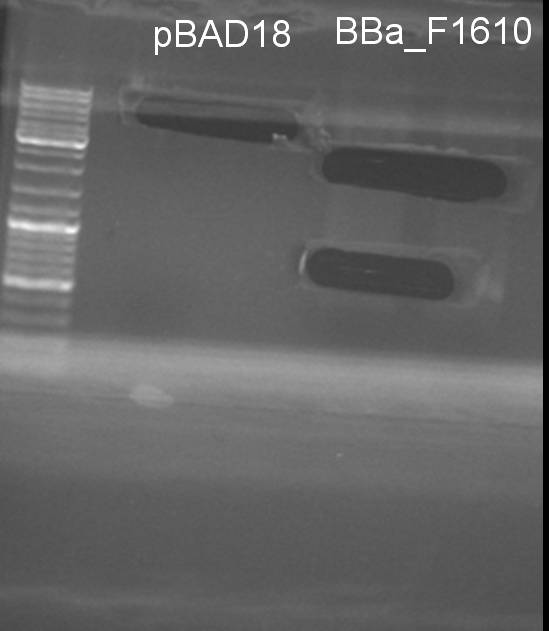
- Miniprep of pBAD18: QIAprep Spin Miniprepkit Kit (250), Qiagen
- Maxiprep of BBa_F1610: CompactPrep Plasmid Mxi Kit 25, Qiagen
- Cloning strategy of pBAD-Sender:
- Amplification of BBa_F1610 with pSB_ins_fw & pSB_ins_rv
- Use XbaI and PstI site to clone into MCS of pBAD18 plasmid
- Digestion of BBa_F1610 and pBAD 18 with PstI and XbaI: Incubation 1.5 h -> 37°C
10.0 µl DNA 3.0 µl NEBuffer 3 3.0 µl BSA 10x 3.0 µl XbaI 2.0 µl PstI 9.0 µl H2O ------- 30.0 µl
- Gelextraction of digestion: Upper band of F1610 seems to be once cutted
- Ligation with insert:vector ratio 3:1: 1h -> RT, Inactivation: 10 min -> 65 °C
6.0 µl Insert 2.0 µl Vector 2.0 µl 10x T4 DNA Ligase Buffer (Fermentas) 1.0 µl T4 DNA Ligase (Fermentas) 9.0 µl H2O ------- 20.0 µl
- Transformation into E. coli TOP 10 cells (Transformation protocol Chris)
- Cloning possibly not works because PstI cuts twice in pBAD18
[back]
Thursday 08/21/2008
Colicin Receiver
Cloning
- Miniprep of pColE1: Miniprepkit, Qiagen
- Inoculation of liquid culture from glycerolstock
Sender part
pBAD-Sender Cloning
- Inoculation of liquidcultures from transformation
General
- Translation of press release into English
- Design of official Wiki
[back]
Friday 08/22/2008
Colicin Receiver
Cloning
- For Colicin-Receiver-Cloning plans see Internal Team Meeting
Activity Test
- For planings see internal team meeting.
Sender part
pBAD-Sender Cloning
- Cloning Results are tested via a activity test
Activity Test
- ONC was used for a sender activity test. The following graph shows the plate scheme for the test. (S = sender, R = Receiver, 1:5 = Sender:Receiver ratio 1:5 ...)
- Cells were incubated and GFP intensity as well as OD were measured every 2 hours. After 2 hours we obtained that we forget to induce LuxI production with arabinose. Because of that 2 µl of a 10% arabinose dilution was putted in every well.
General
- Internal Team Meeting:
- Purification of Colicins:
- Tests on different bacterial strains
- cells vs cells
- supernatant vs cells
- test on eucaryotiv MCF7 and HeLa cells
- Sender Cloning
- Primer design for LuxI (XbaI and HindIII sites)
- While primers have not arrived we use XbaI only for cloning.
- Cloning of Colicin-Receivers
- Purification of pSB1A3 backbone by digestion of BBa_T9002 with Xba I
- Amplification of LuxR Receiver withou GFP from BBa_T9002 with XbaI(fw) and BamHI and SpeI (rv)
- Amplification of colicin operons with BamHI and SpeI restriction sites
- Digestion og LuxR receiver with XbaI and BamHI and colicin operons with BamHI and SpeI
- Ligation of receiver and colicins via BamHI.
- Gelextraction of ligated receiver.
- Ligation of pSB1A3 luxR-colicin-receiver.
- Purification of Colicins:
[back]
 "
"