Team:Heidelberg/Notebook/Killing II/5thweek
From 2008.igem.org
(Difference between revisions)
(→Friday 09/05/2008) |
|||
| (4 intermediate revisions not shown) | |||
| Line 497: | Line 497: | ||
*Preparation of M9 media | *Preparation of M9 media | ||
| - | + | [[https://2008.igem.org/Team:Heidelberg/Notebook/Killing_II/5thweek back]] | |
| - | + | ||
| - | + | ||
==Tuesday 09/02/2008== | ==Tuesday 09/02/2008== | ||
| Line 524: | Line 522: | ||
**New inoculation of same cultures in LB media. | **New inoculation of same cultures in LB media. | ||
| - | + | [[https://2008.igem.org/Team:Heidelberg/Notebook/Killing_II/5thweek back]] | |
| - | + | ||
| - | + | ||
==Wednesday 09/03/2008== | ==Wednesday 09/03/2008== | ||
| Line 564: | Line 560: | ||
prey_Top10,2: 157,4 ng/ul | prey_Top10,2: 157,4 ng/ul | ||
prey_DH5-a,1: 202,0 ng/ul | prey_DH5-a,1: 202,0 ng/ul | ||
| - | prey_DH5-a,2: 174,2 ng/ul | + | prey_DH5-a,2: 174,2 ng/ul |
| - | + | ||
| + | [[https://2008.igem.org/Team:Heidelberg/Notebook/Killing_II/5thweek back]] | ||
==Thursday 09/04/2008== | ==Thursday 09/04/2008== | ||
| Line 621: | Line 617: | ||
*Inoculation of 4 cotransformations for activity tests | *Inoculation of 4 cotransformations for activity tests | ||
| - | + | [[https://2008.igem.org/Team:Heidelberg/Notebook/Killing_II/5thweek back]] | |
==Friday 09/05/2008== | ==Friday 09/05/2008== | ||
| Line 632: | Line 628: | ||
*Activity test over the day: no results | *Activity test over the day: no results | ||
| + | [[https://2008.igem.org/Team:Heidelberg/Notebook/Killing_II/5thweek back]] | ||
[[Team:Heidelberg/Notebook/Killing_II/6thweek | go to 6<sup>th</sup> week]] | [[Team:Heidelberg/Notebook/Killing_II/6thweek | go to 6<sup>th</sup> week]] | ||
Latest revision as of 21:27, 28 October 2008


5th week
Contents |
Monday 09/01/2008
Colicin Receiver
pSB1A3-Receiver-Colicin-Cloning
- Transformationresults of ligation pSB1A3-RecluxpR no colonies
- Retransformation of ligation into TOP 10 -> standard protocoll
Activity Test
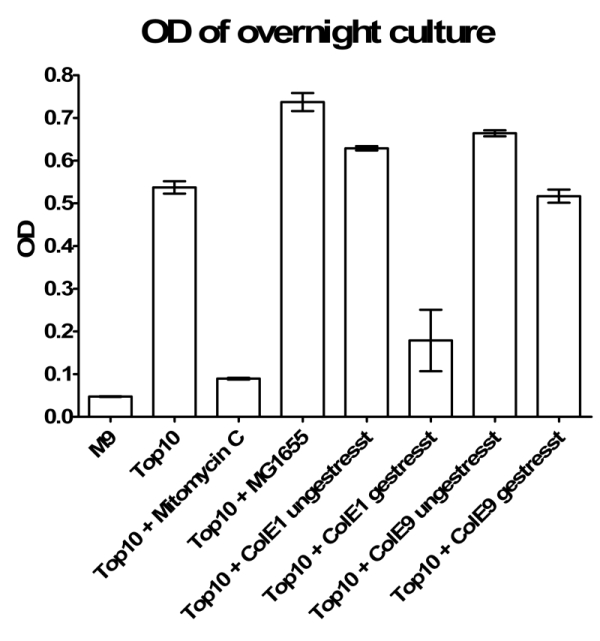
- Inoculation of TOP10 ONC for colicin-test next day:
- +2 ml MG1655
- +0.002 µg/ml Mytomycin C
- 2 ml ColE1 unstressed
- 2 ml ColE1 stressed
- 2 ml ColE9 unstressed
- 2 ml ColE9 stressed
- Inoculation for growthcurve measurement over the day (tomorrow):
- ColE1 unstressed
- ColE1 stressed
- ColE9 unstressed
- ColE9 stressed
- DH5alpha
- TOP10
- MG1655
Sender part
pBAD-Sender Cloning
- Double transformation of pBAD-F1610-9 and AI-2-Sender int TOP 10 and DH5 alphha
- Inoculation of ONC with pBAD-F1610 colony 9 (from glycerolstock)
General
- Preparation of M9 media
[back]
Tuesday 09/02/2008
Colicin Receiver
pSB1A3-Receiver-Colicin-Cloning
- Inoculation of liquid cultures from Transformation of ligation: pSB1A3-RecluxpR.
Activity Test
Sender part
pBAD-Sender Cloning
- Miniprep of pBAD-F1610,9: 179,6 ng/ul; Qiagen Miniprepkit
- Inoculation of liquid cultures from Transformation of double-transformation.
Activity Test
- Results of Colicintest: You can see that stressed Colicin E1 cells are able to kill TOP 10. But this could also be caused by Mytomycin C.
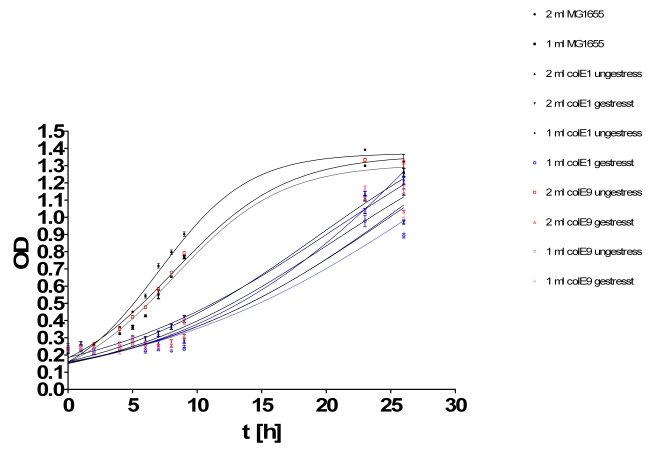
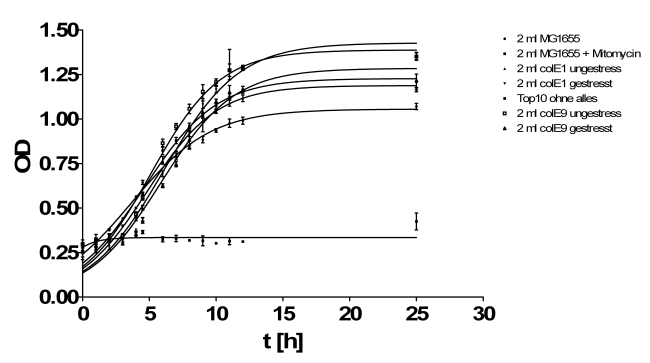
- Growthcurve:
- Inoculation of TOP10s with 1ml/2ml supernatant of:
- MG1655
- ColicinE1 unstressed
- ColicinE1 stressed
- ColicinE9 unstressed
- ColicinE9 stressed
- OD was measured hourly in tecan plate reader
- New inoculation of same cultures in LB media.
- Inoculation of TOP10s with 1ml/2ml supernatant of:
[back]
Wednesday 09/03/2008
Colicin Receiver
pSB1A3-Receiver-Colicin-Cloning
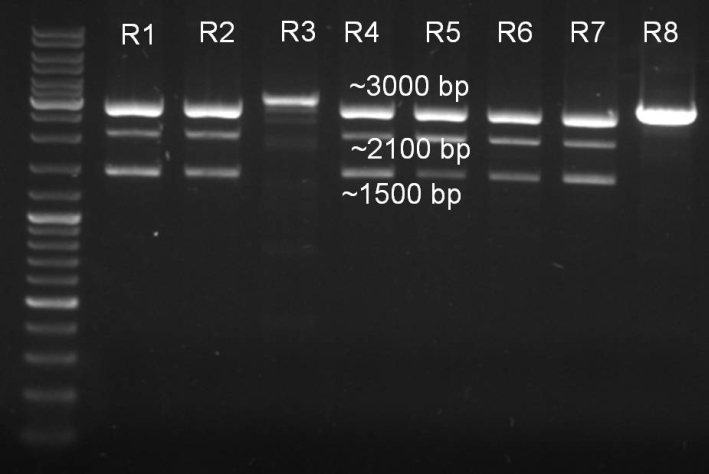
- Minipreps of the ligation ONCsvfrom 02.09.:
concentrations: R1: 152,3 ng/ul R2: 150,4 ng/ul R3: 76,7 ng/ul R4: 132,8 ng/ul R5: 142,1 ng/ul R6: 138,4 ng/ul R7: 136,8 ng/ul R8: 187,7 ng/ul
- Controldigestions of ligation with XbaI and SpeI
10.0 µl DNA 2.0 µl NEBuffer 2 2.0 µl BamHI 2.0 µl XbaI 2.0 µl BSA 10x 2.0 µl H2o ------- 20.0 µl
- Gel of digestion:
- Gelresults: Expected fragments 2150 bp and 1088 bp. The 2150 bp fragments are there but there is a 1500 bp fragment instead of the 1088 bp fragment. Despite this we made some new glycerolstocks.
Activity Test
- Results of growth curve test from yesterday: The results were very diffusing so that we perform this test again.
- New hourly growth curve measurement (see Tuesdaysday for details). No significant effect of colicins can be seen. Only Mytomycin C kills the TOP 10 cells very effectively
Sender part
pBAD-Sender Cloning
- Minipreps of doubletransformations:
concentrations prey_Top10,1: 123,6 ng/ul prey_Top10,2: 157,4 ng/ul prey_DH5-a,1: 202,0 ng/ul prey_DH5-a,2: 174,2 ng/ul
[back]
Thursday 09/04/2008
Colicin Receiver
pSB1A3-Receiver-Colicin-Cloning
- Despite of the strange digestion pattern we try the next step of the cloning.
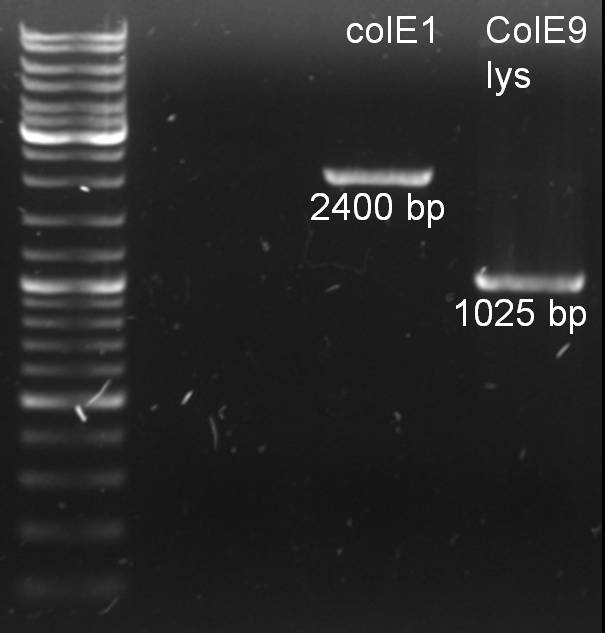
- PCR of colicins:
25.0 µl Phusion MasterMix 2.5 µl colE1prot/colE9prot_fw_BamHI 2.5 µl colE1prot/colE9prot_rv_SpeI 18.0 µl H2O 2.0 µl DNA templates ------- 50.0 µl
program: 98 °C 30 sec 98 °C 10 sec 59 °C 20 sec 72 °C 45 sec 72 °C 8 min 4 °C constant
- PCR purification and analytic gel: Expected fragments are there
- Digestion of purified PCR products with BamHI and SpeI: 1h -> 37 °C
Vector: 6.0 µl DNA 2.0 µl NEBuffer 2 2.0 µl SpeI 2.0 µl BamHI 2.0 µl BSA10x 6.0 µl H2O ------- 20.0 µl
Insert: 10.0 µl DNA 2.0 µl NEBuffer 2 2.0 µl SpeI 2.0 µl BamHI 2.0 µl BSA10x 2.0 µl H2O ------- 20.0 µl
- Gel & Gelextraction: The expected bands were visible. After cutting we purified them using the Qiagen Gel Extraction Kit.
- Sequencing: Send R1 (LuxR-Receiver) to GATC.
- Inoculation of 4 receiver colonies
Activity Test
- New plans for characterization:
- Cloning of Colicin proteins into pQE-30 vector for His-Tag purification of colicins. Therefore we designed reverse primers with HindIII (ColE1) and XmaI(ColE9) sites.
Sender part
pBAD-Sender Cloning
- Inoculation of 4 cotransformations for activity tests
[back]
Friday 09/05/2008
Colicin Receiver
pSB1A3-Receiver-Colicin-Cloning
- Ligation and Transformation of pRecluxpR with ColE1/ColE9: no results
Sender part
Activity Test
- Activity test over the day: no results
[back]
 "
"