Team:Harvard/Shewie
From 2008.igem.org
| Line 65: | Line 65: | ||
==Molecular Biology with <i>Shewanella oneidensis</i>== | ==Molecular Biology with <i>Shewanella oneidensis</i>== | ||
| - | {|align="justify" style="background-color:#FFFFFF;text-indent: | + | {|align="justify" style="background-color:#FFFFFF;text-indent: 15pt; text-align:justify" width="100%" |
| | | | ||
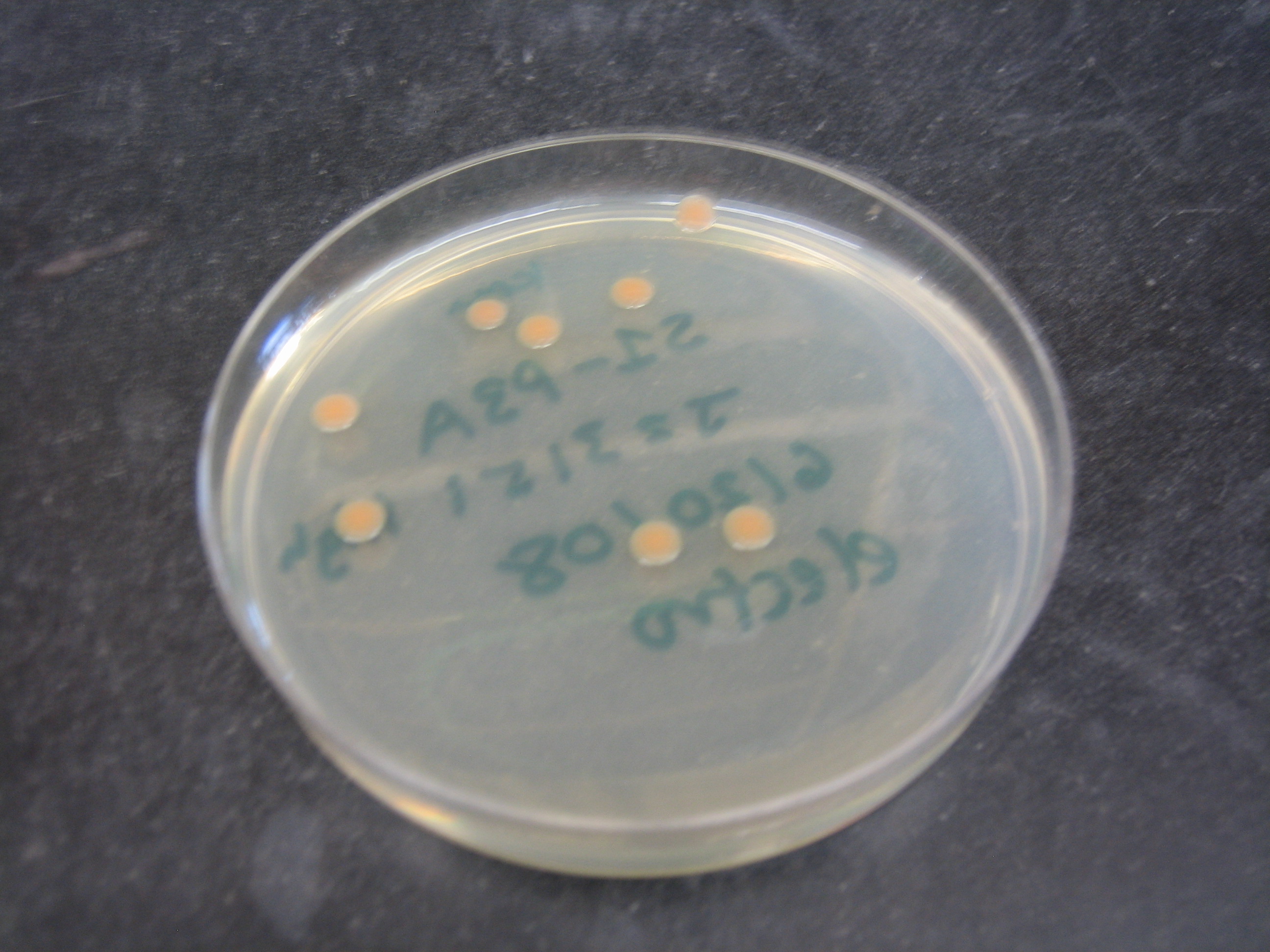
| - | + | The ''Shewanella oneidensis MR-1'' genome was sequenced in 2002, greatly increasing its usefulness as a model organism. It was found that it had a 4,969,803 base pair circular chromosome and a 161,613 base pair plasmid (Heidelberg et al. 2002). When cloning in ''S. oneidensis MR-1'', it has also been shown that plasmids with p15A origins replicate freely, whereas plasmids with a pMB1 origin of replication do not (Myers and Myers 1997). [We further found that …]. ''S. oneidensis MR-1'' grows at 30 ºC, can be electroporated (see protocol in our [[Team:Harvard/GenProtocols| Notebook]]) and forms round orange pink colonies on plates. It is resistant to ampicillin, but other resistance markers work (Daad). Together, these characteristics make ''S. oneidensis MR-1'' a genetically tractable organism good for exploring the possibilities of regulated bacterial electrical output.</p> | |
|[[Image:Shewanella colonies growing on plate.JPG|thumb|300px|''S. oneidensis MR-1'' colonies from a transformation]] | |[[Image:Shewanella colonies growing on plate.JPG|thumb|300px|''S. oneidensis MR-1'' colonies from a transformation]] | ||
|- | |- | ||
Revision as of 01:59, 29 October 2008
|
|
 "
"