Team:Heidelberg/Notebook/Killing I/Notebook/week12
From 2008.igem.org
(→Monday, 10/20/08) |
|||
| Line 489: | Line 489: | ||
* lane 9: old insert fully mutated | * lane 9: old insert fully mutated | ||
* results: the new minipreps seem to be the old unmutated pBluescript | * results: the new minipreps seem to be the old unmutated pBluescript | ||
| + | |||
| + | |||
| + | ===Characterization of oriT=== | ||
| + | *Quantitatively test for oriT <br> | ||
| + | Donor: overnight culture Top10 oriT+pUB307 OD(600nm): 1.548 <br> | ||
| + | Recipient: <br> | ||
| + | overnight culture Top10 pBAD 33 OD(600nm): 1.808 <br> | ||
| + | overnight culture MG1655 pBAD 33 OD(600nm): 2.932<br> | ||
| + | overnight culture DH5alpha pBAD 33 OD(600nm): 1.976 <br> | ||
| + | **Centrifuge overnight culture in 1.5ml eppi for 2min at 13000rpm, 24 samples donor, 510ul/sample; 8samples Top10 recipient, 450ul/sample; 8samples MG1655 recipient, 375ul/sample; 8samples DH5alpha recipient, 600ul/sample; | ||
| + | **Wash the pellet twice with LB medium | ||
| + | **Resolve the pellet in LB medium | ||
| + | **Centrifuge the washed recipient for 2min at 13000rpm, discard the fluid | ||
| + | **Add the washed donor suspension | ||
| + | **Vortex and resolve the pellet | ||
| + | **Centrifuge the mix for 1min at 13000rpm | ||
| + | **Resolve the pellet in 100ul LB | ||
| + | **Put membrane filter on the LB agar | ||
| + | **Pipett the suspension on membrane filter (8samples*3tests) | ||
| + | **Incubate the plates with membrane filter at 37°C | ||
| + | **Put directly one membrane filter into 1ml LB in an 1.5ml eppi | ||
| + | **Vortex the eppi for 30sec, dilute for 10-5 and 10-6, plate them out on LB/Amp+Cm plates (0min) | ||
| + | **For Top10 as recipient: plate them out also on LB/Kan+Cm plates and LB/Amp+Kan+Cm plates | ||
| + | **After 6, 12, 18, 24, 30, 36, 42 min repeat the last three steps. | ||
| + | **Negative control plates: | ||
| + | ***LB/Cm+Amp: <br> | ||
| + | 100ul donor overnight culture<br> | ||
| + | 100ul recipient overnight culture (x3) | ||
| + | **Cell number determination | ||
| + | ***LB/Cm: 100ul 10-6 recipient overnight culture (x3) | ||
| + | ***LB/Kan+Amp: 100ul 10-6 donor overnight culture | ||
| + | <br> | ||
| + | *Result: | ||
| + | **Negative control: negative | ||
| + | **Colony on LB/Cm: <br> | ||
| + | Top10:166 (Titer of recipient: 1.66e9/ml) <br> | ||
| + | MG1655:194 (Titer of recipient: 1.94e9/ml) <br> | ||
| + | DH5alpha:115 (Titer of recipient: 1.15e9/ml) <br> | ||
| + | **Colony on LB/Kan+Amp: 140 (Titer of donor: 1.4e9/ml)<br> | ||
| + | (information for volume counting: 1OD Top10 = 9e8/ml; 1OD MG1655 = 6.6e8/ml; 1OD DH5alpha = 5.8e8/ml) | ||
| + | **Colony on other LB/Cm+Amp plates, LB/Kan+Cm plates and LB/Amp+Kan+Cm plates: | ||
| + | ***10-5 dilute: <br> | ||
| + | Time Top10-CA Top10-CK Top10-CKA MG1655 DH5alpha<br> | ||
| + | 0 37 2 1 0 7<br> | ||
| + | 6 390 17 4 1 22<br> | ||
| + | 12 1390x 194 90 7 480<br> | ||
| + | 18 1720x 230 109 49 1180x<br> | ||
| + | 24 2880x 1130x 800x 136 1920x<br> | ||
| + | 30 3500x 2500x 1540x 260 2960x<br> | ||
| + | 36 3750x 2600x 1800x 920x 3440x<br> | ||
| + | 42 960x 4040x<br> | ||
| + | <br>x:10-6 dilute | ||
==Tuesday, 10/21/08== | ==Tuesday, 10/21/08== | ||
Revision as of 13:33, 29 October 2008


| << Week 11 | Overview | Week 13 >> |
|---|
Week 12
Contents |
Monday, 10/20/08
Proceedings of phage cloning strategy one
- the fully mutated insert in pBluescript SK II was digested with XbaI/XhoI and the insert extracted from the gel
- lambda DNA was also cut with XbaI/XhoI and the backbone also extracted from the gel.
- the insert was ligated in the lambda backbone over night using standard ligation methods
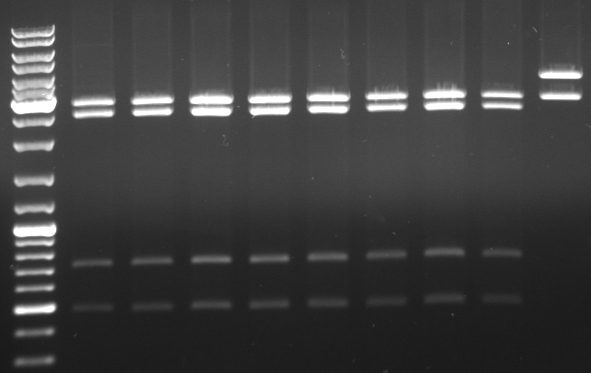
Phage cloning strategy two
- Digestion with XbaI/XhoI
- should be: 3945, 2898
- Gel
- lane 1-8: new minipreps
- lane 9: old insert fully mutated
- results: the new minipreps seem to be the old unmutated pBluescript
Characterization of oriT
- Quantitatively test for oriT
Donor: overnight culture Top10 oriT+pUB307 OD(600nm): 1.548
Recipient:
overnight culture Top10 pBAD 33 OD(600nm): 1.808
overnight culture MG1655 pBAD 33 OD(600nm): 2.932
overnight culture DH5alpha pBAD 33 OD(600nm): 1.976
- Centrifuge overnight culture in 1.5ml eppi for 2min at 13000rpm, 24 samples donor, 510ul/sample; 8samples Top10 recipient, 450ul/sample; 8samples MG1655 recipient, 375ul/sample; 8samples DH5alpha recipient, 600ul/sample;
- Wash the pellet twice with LB medium
- Resolve the pellet in LB medium
- Centrifuge the washed recipient for 2min at 13000rpm, discard the fluid
- Add the washed donor suspension
- Vortex and resolve the pellet
- Centrifuge the mix for 1min at 13000rpm
- Resolve the pellet in 100ul LB
- Put membrane filter on the LB agar
- Pipett the suspension on membrane filter (8samples*3tests)
- Incubate the plates with membrane filter at 37°C
- Put directly one membrane filter into 1ml LB in an 1.5ml eppi
- Vortex the eppi for 30sec, dilute for 10-5 and 10-6, plate them out on LB/Amp+Cm plates (0min)
- For Top10 as recipient: plate them out also on LB/Kan+Cm plates and LB/Amp+Kan+Cm plates
- After 6, 12, 18, 24, 30, 36, 42 min repeat the last three steps.
- Negative control plates:
- LB/Cm+Amp:
- LB/Cm+Amp:
100ul donor overnight culture
100ul recipient overnight culture (x3)
- Cell number determination
- LB/Cm: 100ul 10-6 recipient overnight culture (x3)
- LB/Kan+Amp: 100ul 10-6 donor overnight culture
- Cell number determination
- Result:
- Negative control: negative
- Colony on LB/Cm:
Top10:166 (Titer of recipient: 1.66e9/ml)
MG1655:194 (Titer of recipient: 1.94e9/ml)
DH5alpha:115 (Titer of recipient: 1.15e9/ml)
- Colony on LB/Kan+Amp: 140 (Titer of donor: 1.4e9/ml)
- Colony on LB/Kan+Amp: 140 (Titer of donor: 1.4e9/ml)
(information for volume counting: 1OD Top10 = 9e8/ml; 1OD MG1655 = 6.6e8/ml; 1OD DH5alpha = 5.8e8/ml)
- Colony on other LB/Cm+Amp plates, LB/Kan+Cm plates and LB/Amp+Kan+Cm plates:
- 10-5 dilute:
- 10-5 dilute:
- Colony on other LB/Cm+Amp plates, LB/Kan+Cm plates and LB/Amp+Kan+Cm plates:
Time Top10-CA Top10-CK Top10-CKA MG1655 DH5alpha
0 37 2 1 0 7
6 390 17 4 1 22
12 1390x 194 90 7 480
18 1720x 230 109 49 1180x
24 2880x 1130x 800x 136 1920x
30 3500x 2500x 1540x 260 2960x
36 3750x 2600x 1800x 920x 3440x
42 960x 4040x
x:10-6 dilute
Tuesday, 10/21/08
Proceedings of phage cloning strategy one
- the emerging plasmid consisting of our insert in tha lambda phage was transformed in E. coli by chemical transformation and in vitro packaging
- the bacterias were plated on chloramphenicol and incubated at 28 °C to ensure that the thermolabile cI (cI857) was not degraded
Phage cloning strategy two
- do again a digestion of mutagenesis pcr minipreps from 10/07/08 but with an additional restriction enzyme (EcoRI)to get a better separation on gel to cut out the backbone
- digestion with KpnI/AgeI/EcoRI
- mutagenesis pcr succesful: 300bp, 1.4kb 5.1kb
- mutagenesis pcr not succesful: 300bp, 1.4kb, 1.7kb, 3.4kb
- -->mutagenesis pcr on the first three lanes successful
- cut out 5.1kb band and gel purification kit
- lane0: DNA ladder mix
- lane1: purified 5.1kb band from last gel (pBluescript backbone from phage cloning strategy one (KpnI/AgeI)
- lane2: CmR1 digested (KpnI/SacI)
- lane3: CmR2 digested (KpnI/SacI)
- lane4: GFP1 digested (SacI/AgeI)
- lane5: GFP2 digested (SacI/AgeI)
- ligation of purified 5.1kb band from last gel (pBluescript backbone from phage cloning strategy one (KpnI/AgeI) with CmR1/2 and GFP1/2
- 30min at room temperature
- transformation in TOP10
Wednesday, 10/22/08
Proceedings of phage cloning strategy one
- first small colonies can be seen on chloramphenicol plates.
- colonies were picked and transferred in liquid media containing chloramphenicol
Thursday, 10/23/08
Proceedings of phage cloning strategy one
- colonies could grow in chloramphenicol containing medium --> samll aliquots were transferred in ampicillin containing medium to check for the resisitance
- colony PCRs were conducted to ensure that our insert is embedded in the growing cells
- pUB307 was transferred in cells harboring our modified lambda phage by conjugation --> plating on Cm + Kan
phage cloning strategy two
- inoculation of liquid cultures from ligation (tuesday)
Friday, 10/24/08
Proceedings of phage cloning strategy one
- colonies grew on plates containing chloramphenicol and kanamycin --> were picked and liquid medium inoculated
- conjugation of our lambda phage into cells harboring T9002 in pSB1A3 --> brought out on plates containing chloramphenicol and ampicillin
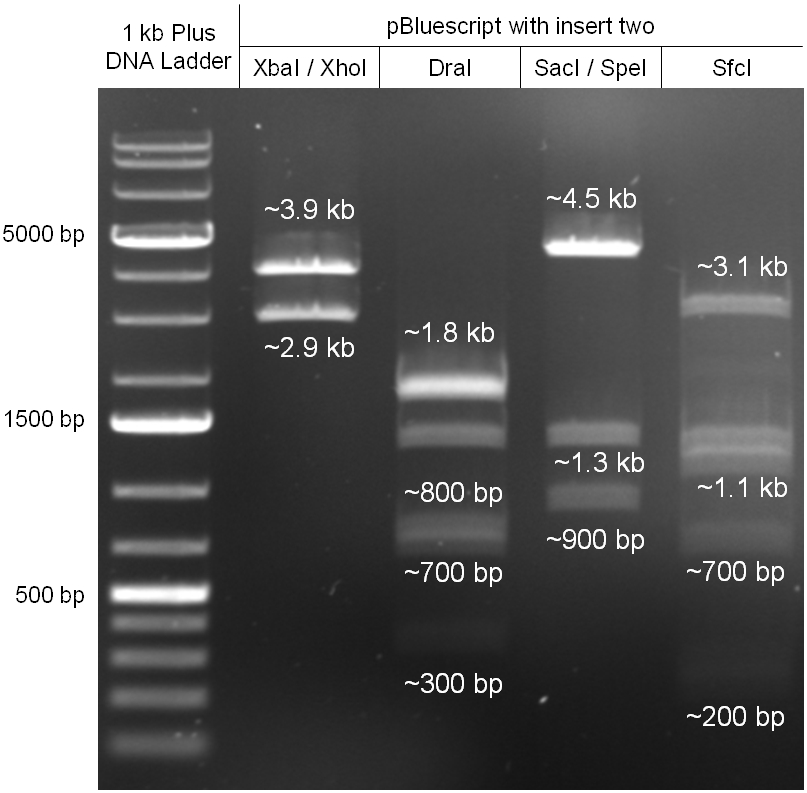
phage cloning strategy two
- Miniprep of overnight cultures
- 4 Control digestions to check wheter the ligation was successful or not
- lane0: DNA ladder mix
- lane1: XbaI/XhoI: 3,9kb / 2,9kb (NEB2, BSA)
- lane2: DraI: 1,85kb / 1,8kb / 1,35kb / 780bp / 692bp/ 339bp/ 19bp (NEB4)
- lane3: SacI/SpeI: 4.5kb / 1.3kb / 929bp / 34bp (NEB1,BSA)
- lane4: SfcI: 3.1kb / 1.3kb / 1.1kb / 678bp / 247bp / 192bp (NEB4, BSA)
- we got the correct pBluescript with the insert of phage cloning strategy two
Saturday, 10/25/08
Proceedings of phage cloning strategy one
- we got colonies on the plates with ampicillin and chloramphenicol only when our conjugated cultures were brought out, but not if T9002 or the cells harboring pUB307 and our phage were brought out separately
- this shows that our phage can be transported by conjugation
Sunday, 10/26/08
Chloramphenicol resistance cassette
- Experiment, to check until what antibiotic concentration the plasmid gives resistance
- inoculation of TOP10 and TOP10 with BBa_K150003 in LB with different Cm concentrations:
- 0 µg/ml
- 5 µg/ml
- 10 µg/ml
- 15 µg/ml
- 20 µg/ml
- 25 µg/ml
- 30 µg/ml
- 35 µg/ml
- 40 µg/ml
- 45 µg/ml
- 50 µg/ml
- 60 µg/ml
- 70 µg/ml
- 80 µg/ml
- 90 µg/ml
- 100 µg/ml
- 250 µg/ml
- 500 µg/ml
- 750 µg/ml
- 1000 µg/ml
- 5000 µg/ml
- stock solution: 100mg/ml in ethanol
- mock treated:
- 500 µg/ml
- 750 µg/ml
- 1000 µg/ml
- 5000 µg/ml
- meausurement of OD every 30min with TECAN, measurement length: 12 hours
| << Week 11 | Overview | Week 13 >> |
|---|
 "
"