Team:Rice University/Notebook/20 June 2008
From 2008.igem.org
(Difference between revisions)
StevensonT (Talk | contribs) |
StevensonT (Talk | contribs) |
||
| (7 intermediate revisions not shown) | |||
| Line 6: | Line 6: | ||
*#Infected culture was heat shocked @ 42*C for 20min, inducing lysis by denaturing the CI857 repressor. | *#Infected culture was heat shocked @ 42*C for 20min, inducing lysis by denaturing the CI857 repressor. | ||
*#Cells were then grown and their optical density was measured at different time points after heat shock. | *#Cells were then grown and their optical density was measured at different time points after heat shock. | ||
| + | *#'''Result'''-lysis was never observed. Cells may need to be maintained at higher temperature to ensure all phage infections result in lysis. | ||
**Lysogeny Screen | **Lysogeny Screen | ||
| - | *# '''Result | + | *# '''Result'''-An infected VCS257 culture was streaked onto LB-agar. Isolated colonies were used to innoculate a 96 deep-well plate which was grown O/N @ 30*C shaking @ 300rpm. Using the well replicator, each of the wells was spotted onto two large petri dishes. One dish was grown @ 30*C and the other was grown @ 42*C O/N. Spots that grew on 30*C plates and not on 42*C plates are lysogens as the phage infected cells would lyse due to the temperature sensitive cI. The plates show that there is only one colony that could be a possible lysogen. |
*# The possible lysogen colony was streaked onto LB plates to be grown O/N @ 30*C and 42*C to verify temperature screen. | *# The possible lysogen colony was streaked onto LB plates to be grown O/N @ 30*C and 42*C to verify temperature screen. | ||
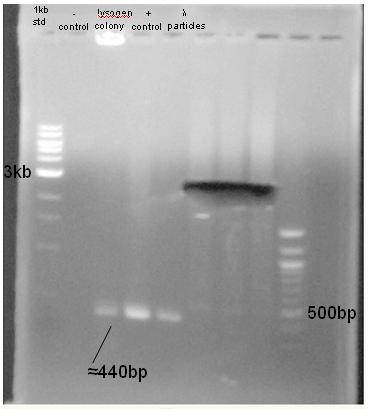
*# A colony PCR was performed on the possible lysogen using the CroF and CroR primers. If a copy of the lambda genome is incorporated into the lysogen's, the PCR should result in amplification of the phage gene ''cro'' and produce a 400bp product. Additionally, three other PCR reactions were performed in parallel. | *# A colony PCR was performed on the possible lysogen using the CroF and CroR primers. If a copy of the lambda genome is incorporated into the lysogen's, the PCR should result in amplification of the phage gene ''cro'' and produce a 400bp product. Additionally, three other PCR reactions were performed in parallel. | ||
| Line 13: | Line 14: | ||
*## -positive control using lambda DNA | *## -positive control using lambda DNA | ||
*## -second sample using lambda particles from stocks | *## -second sample using lambda particles from stocks | ||
| + | *#'''Result'''-PCR gel analysis shows that the bacterial colony, phage DNA, and lambda particles all produce a PCR product at approximately 440bp, where the amplified ''cro'' should be. This data further supports the conclusion that the screened colony is a lysogen, and that it is possible to use packaged phage DNA as a template in a PCR. | ||
| + | *#'''Result'''-Temperature screen of the colony showed that streaked plates did not grow colonies @ 42*C and grew an abundant number @ 30*C. | ||
| + | [[Image:lysogeny screen.jpg|400px]] | ||
| + | <BR><BR><BR><BR> | ||
| + | {| style="color:#1b2c8a;background-color:#0c6;" cellpadding="3" cellspacing="1" border="1" bordercolor="#fff" width="62%" align="center" | ||
| + | !align="center"|[[Team:Rice_University|Home]] | ||
| + | !align="center"|[[Team:Rice_University/Team|The Team]] | ||
| + | !align="center"|[[Team:Rice_University/Project|The Project]] | ||
| + | !align="center"|[[Team:Rice_University/Parts|Parts Submitted to the Registry]] | ||
| + | !align="center"|[[Team:Rice_University/Modeling|Modeling]] | ||
| + | !align="center"|[[Team:Rice_University/Notebook|Notebook]] | ||
| + | |} | ||
Latest revision as of 20:27, 25 June 2008
Friday, 20 June
- Taylor Stevenson:
- Lysis Assay of VCS257 SupA+
- Cells were grown O/N @ 37*C in Lambda media.
- 4.5mL culture was infected with 4.5uL of phage packaging product from November, 2007 and allowed to incubate for 20min @ 37*C shaking @ 250 rpm.
- Infected culture was heat shocked @ 42*C for 20min, inducing lysis by denaturing the CI857 repressor.
- Cells were then grown and their optical density was measured at different time points after heat shock.
- Result-lysis was never observed. Cells may need to be maintained at higher temperature to ensure all phage infections result in lysis.
- Lysogeny Screen
- Result-An infected VCS257 culture was streaked onto LB-agar. Isolated colonies were used to innoculate a 96 deep-well plate which was grown O/N @ 30*C shaking @ 300rpm. Using the well replicator, each of the wells was spotted onto two large petri dishes. One dish was grown @ 30*C and the other was grown @ 42*C O/N. Spots that grew on 30*C plates and not on 42*C plates are lysogens as the phage infected cells would lyse due to the temperature sensitive cI. The plates show that there is only one colony that could be a possible lysogen.
- The possible lysogen colony was streaked onto LB plates to be grown O/N @ 30*C and 42*C to verify temperature screen.
- A colony PCR was performed on the possible lysogen using the CroF and CroR primers. If a copy of the lambda genome is incorporated into the lysogen's, the PCR should result in amplification of the phage gene cro and produce a 400bp product. Additionally, three other PCR reactions were performed in parallel.
- -negative control using water
- -positive control using lambda DNA
- -second sample using lambda particles from stocks
- Result-PCR gel analysis shows that the bacterial colony, phage DNA, and lambda particles all produce a PCR product at approximately 440bp, where the amplified cro should be. This data further supports the conclusion that the screened colony is a lysogen, and that it is possible to use packaged phage DNA as a template in a PCR.
- Result-Temperature screen of the colony showed that streaked plates did not grow colonies @ 42*C and grew an abundant number @ 30*C.
| Home | The Team | The Project | Parts Submitted to the Registry | Modeling | Notebook |
|---|
 "
"